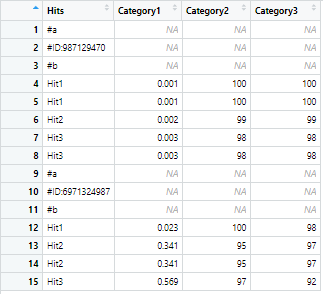
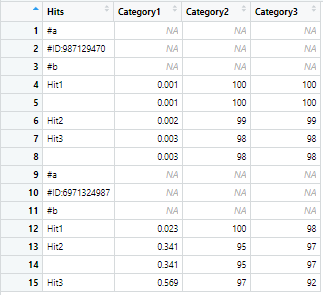
I have a test dataframe df from which I want to drop duplicate values in the Hits column but not the rows associated with the duplicate values. The condition however is that the dropping is to be done only in some specific ranges of row indices.
df <- data.frame(
Hits = c("#a", "#ID:987129470", "#b", "Hit1", "Hit1", "Hit2", "Hit3", "Hit3", "#a", "#ID:6971324987", "#b", "Hit1", "Hit2", "Hit2", "Hit3"),
Category1 = c(NA, NA, NA, 0.001, 0.001, 0.002, 0.003, 0.003, NA, NA, NA, 0.023, 0.341, 0.341, 0.569),
Category2 = c(NA, NA, NA, 100, 100, 99, 98, 98, NA, NA, NA, 100, 95, 95, 97),
Category3 = c(NA, NA, NA, 100, 100, 99, 98, 98, NA, NA, NA, 98, 97, 97, 92))
df looks like this
The ranges of row indices in which the dropping operation is to be carried out are 4:8 and 12:15 in this case. Basically, the duplicate hits under each ID are to be dropped, keeping the associated values in the other columns intact. The output should look something like this
In the original dataframe (which has ~100k rows!) it is not possible to specify the ranges. How do I solve this issue?
CodePudding user response:
First make groups by starting with #a
then use an ifelse statement.
library(dplyr)
df %>%
group_by(id_Group = cumsum(Hits=="#a")) %>%
mutate(Hits = ifelse(duplicated(Hits), "", Hits)) %>%
ungroup() %>%
select(-id_Group)
Hits Category1 Category2 Category3
<chr> <dbl> <dbl> <dbl>
1 "#a" NA NA NA
2 "#ID:987129470" NA NA NA
3 "#b" NA NA NA
4 "Hit1" 0.001 100 100
5 "" 0.001 100 100
6 "Hit2" 0.002 99 99
7 "Hit3" 0.003 98 98
8 "" 0.003 98 98
9 "#a" NA NA NA
10 "#ID:6971324987" NA NA NA
11 "#b" NA NA NA
12 "Hit1" 0.023 100 98
13 "Hit2" 0.341 95 97
14 "" 0.341 95 97
15 "Hit3" 0.569 97 92
CodePudding user response:
Another possible solution:
library(tidyverse)
df <- data.frame(
Hits = c("#a", "#ID:987129470", "#b", "Hit1", "Hit1", "Hit2", "Hit3", "Hit3", "#a", "#ID:6971324987", "#b", "Hit1", "Hit2", "Hit2", "Hit3"),
Category1 = c(NA, NA, NA, 0.001, 0.001, 0.002, 0.003, 0.003, NA, NA, NA, 0.023, 0.341, 0.341, 0.569),
Category2 = c(NA, NA, NA, 100, 100, 99, 98, 98, NA, NA, NA, 100, 95, 95, 97),
Category3 = c(NA, NA, NA, 100, 100, 99, 98, 98, NA, NA, NA, 98, 97, 97, 92))
df %>%
group_by(Hits, if_else(str_detect(Hits, "Hits*"), 1, 0) %>% data.table::rleid(.)) %>%
mutate(Hits = if_else(row_number() > 1 & str_detect(Hits, "Hits*"), "", Hits)) %>%
ungroup %>% select(-last_col())
#> # A tibble: 15 × 4
#> Hits Category1 Category2 Category3
#> <chr> <dbl> <dbl> <dbl>
#> 1 "#a" NA NA NA
#> 2 "#ID:987129470" NA NA NA
#> 3 "#b" NA NA NA
#> 4 "Hit1" 0.001 100 100
#> 5 "" 0.001 100 100
#> 6 "Hit2" 0.002 99 99
#> 7 "Hit3" 0.003 98 98
#> 8 "" 0.003 98 98
#> 9 "#a" NA NA NA
#> 10 "#ID:6971324987" NA NA NA
#> 11 "#b" NA NA NA
#> 12 "Hit1" 0.023 100 98
#> 13 "Hit2" 0.341 95 97
#> 14 "" 0.341 95 97
#> 15 "Hit3" 0.569 97 92