I am putting together a workflow in R that will ultimately be used to assist in migrating a series of very large databases that are similar, but frustratingly different in minor ways.
One of the things I need to be able to visualise is which variable names are present in each database, and what datatype they are.
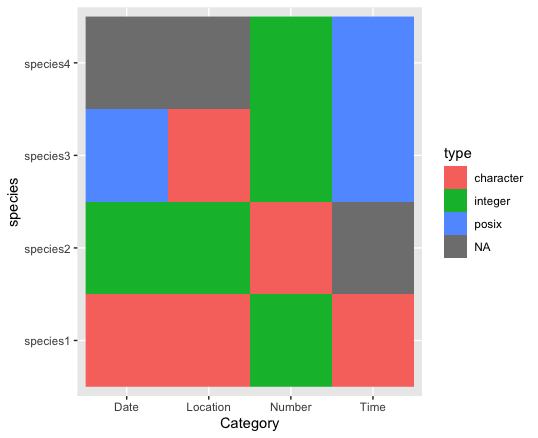
I have reached the point where I have a summary dataframe that looks very similar to the example below.
category <- c("Location", "Date", "Time", "Number")
species1 <- c("character", "character", "character", "integer")
species2 <- c("integer", "integer", NA, "character")
species3 <- c("character", "posix", "posix", "integer")
species4 <- c(NA, NA, "posix", "integer")
comparison_table <- data.frame(category, species1, species2, species3, species4)
The NA values denote that this variable is not present within a specific database.
My ultimate goal was to construct a plot of coloured squares to easily identify inconsistent datatypes between the databases (for example, where dates have been recorded as integers instead of POSIX, or where latitude recorded as a character instead of an integer).
My gut tells me that the geom_raster in ggplot2 should be the simplest way to achieve this, but I keep coming up short. I know that I need to define the fill in the aesthetic, but every attempt is met with a different error.
comparison_table %>%
ggplot(aes(x = colnames(comparison_table), y = rownames(comparison_table)))
geom_raster()
A fresh pair of eyes and a less tired brain would be deeply appreciated.
CodePudding user response:
You'll need to re-structure your data to fit in the grammar of ggplot.
In aes(x, y, fill), we should supply a column from data, which essentially tells which variable should the subsequent geom_function() use to display the data.
In your case, you want:
- x-asix to be "Location", "Date", "Time", "Number", which I grouped them into the
Categorycolumn. - Similarly, your y-axis would be
species, where species1, species2, species3, species4 are grouped in this column, with it's corresponding value grouped into thetypecolumn. - Moreover, since you wish to visualise your data using
geom_raster(), you should also tellggplotto use which variable tofillthe squares (remember to usefillinsideaes()if yourfillcomes from a column).
library(tidyverse)
comparison_table %>% pivot_longer(!Category, names_to = "species", values_to = "type") %>%
ggplot(aes(x = Category, y = species, fill = type))
geom_raster()