What I'm trying to do: plot spatial objects with ggspatial::geom_sf(), using coord_sf() or a combination of ggspatial::layer_spatial() and ggspatial::annotation_spatial() to specify the extent of the of the plot.
This ggplot/ggspatial behavior has been described in several posts, but the "solutions" have been just ad-hoc hacks that do nothing to ensure the issue doesn't re-occur. See:
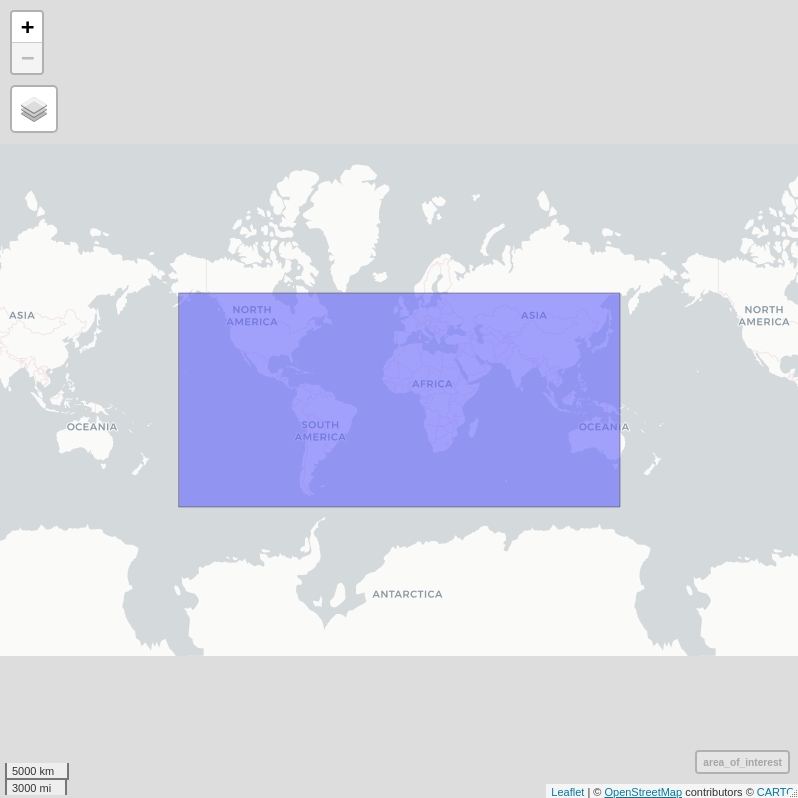
area_of_interest <- matrix(c(-160, -60,
150, -60,
150, 60,
-160, 60,
-160, -60),
byrow = TRUE,
ncol = 2) %>%
list() %>%
st_polygon() %>%
st_sfc(crs = 4326)
mapview(area_of_interest) # this is our area of interest
# an atribute of points - in AOI or not?
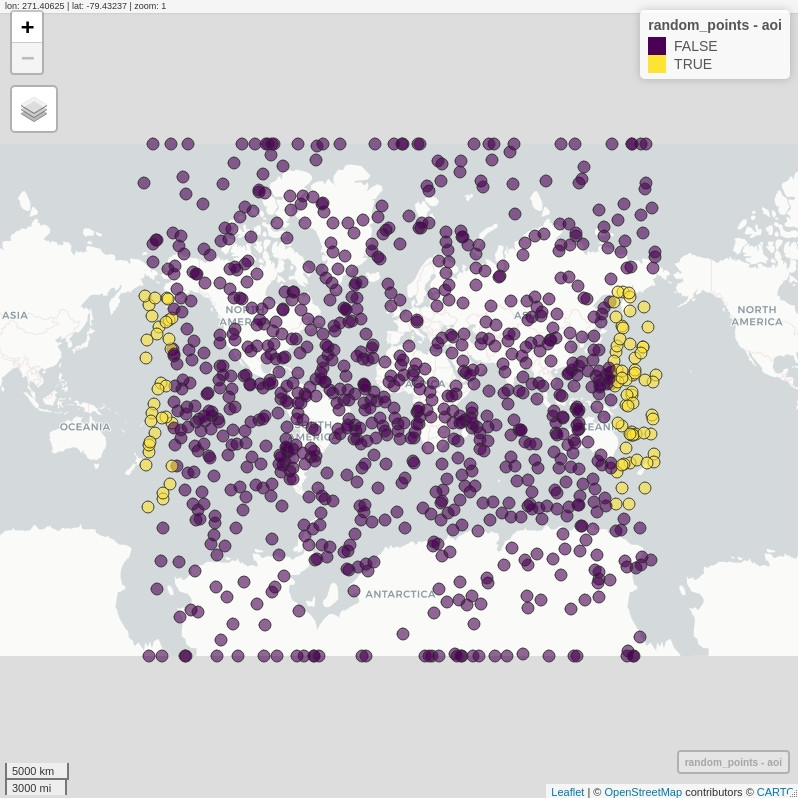
random_points$aoi <- st_contains(area_of_interest,
random_points,
sparse = F) %>%
t() %>%
c()
# a visual overview; this is _not_ expected!
# the topology of AOI was not applied correctly
mapview(random_points, zcol = "aoi")
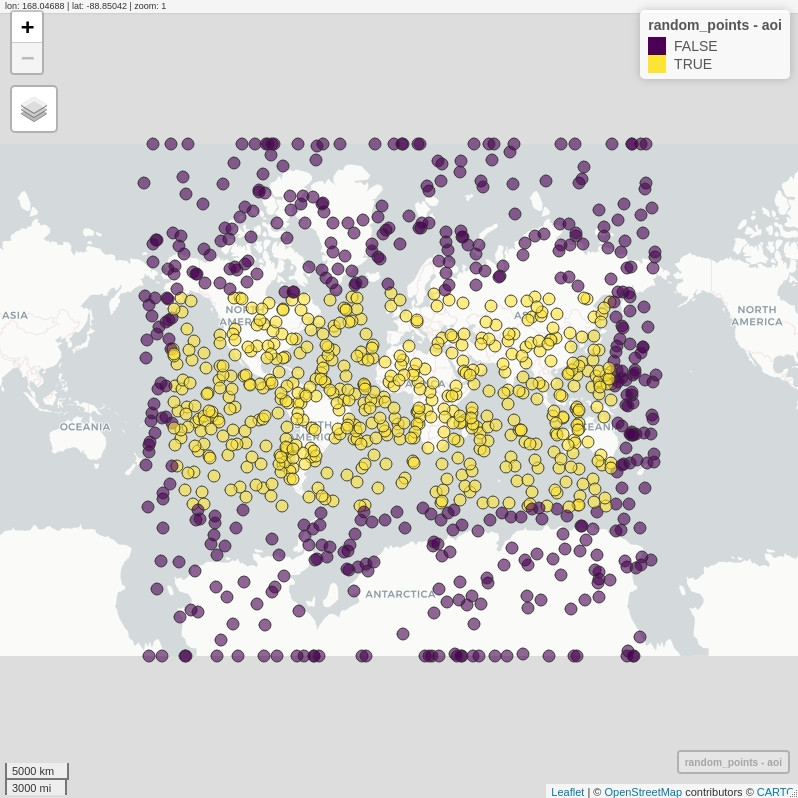
# let us try turning S2 engine off, and fall back to good old GEOS
sf_use_s2(F)
# exactly the same code as before!!
random_points$aoi <- st_contains(area_of_interest,
random_points,
sparse = F) %>%
t() %>%
c()
# but the output behaves much better!
mapview(random_points, zcol = "aoi")
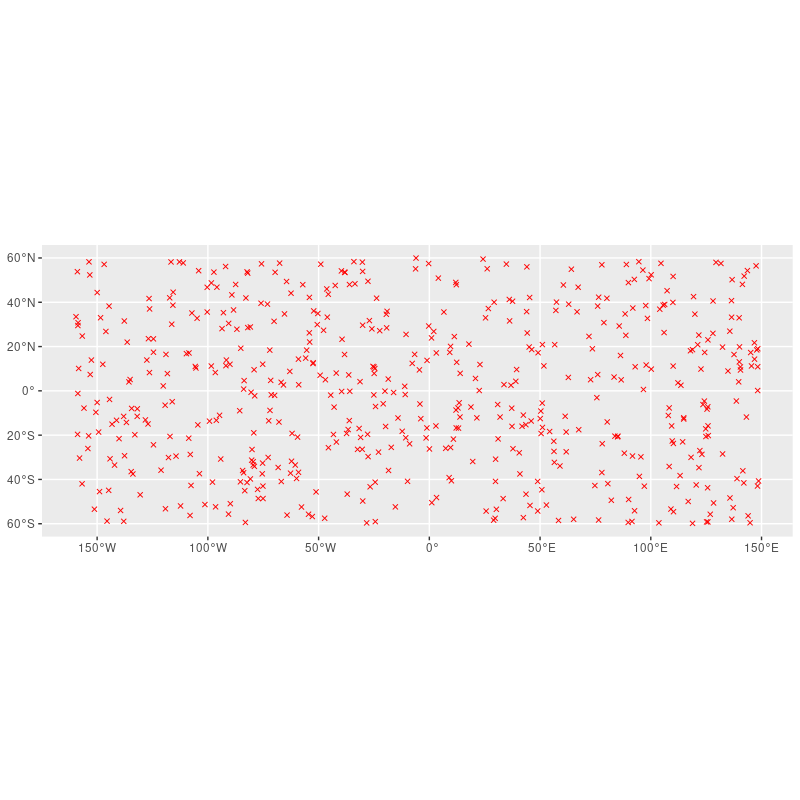
# a seletion of random points; cropped to area of interest
library(ggplot2)
random_points %>%
filter(aoi) %>%
ggplot() geom_sf(pch = 4, color = "red")