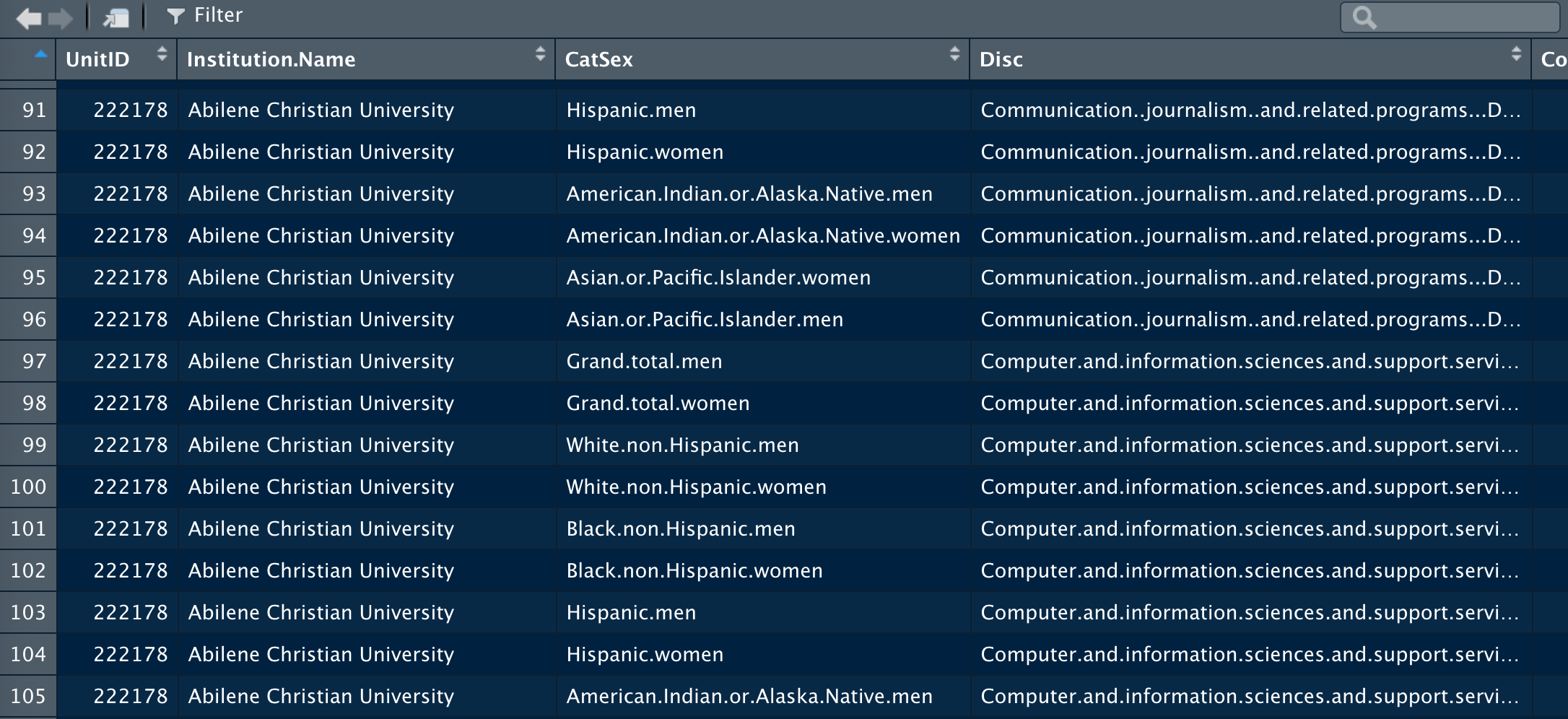
I have a dataset with a column, CatSex, that's got data in it in a form similar to "American.Indian.or.Alaska.Native.men"--the characters after the last period, I want to turn into a new pivoted column, so I have two columns, one called Cat with only the demographic info in it, and one called Sex with the sex in it. The characters before the sex designation don't follow any clear pattern. I am not very good at R, but it's better than Tableau Prep with large data sets, it seems. What I ultimately want is to pivot the data so that I have two distinct columns for the different categories here. I used this code to get part of the way there (the original data held like 119 columns with names like "Grand.total.men..C2005_A_RV..First.major..Area..ethnic..cultural..and.gender.studies...Degrees.total"), but I can't figure out how to do this with the pattern I'm now left with in the column CatSex:
pivot_longer(
cols = -c(UnitID, Institution.Name),
names_to = c("CatSex", "Disc"),
names_pattern = "(.*)..C2005_A_RV..First.major..(.*)",
values_to = "Count",
values_drop_na = TRUE
)
Here's a screenshot of the data structure I have now. I'm sorry for not putting in reproducible code--I don't know how to do that in this context!
EDIT: Here's a head(df) of the cleaned data so far:
# A tibble: 6 × 5
UnitID Institution.Name CatSex Disc Count
<int> <fct> <chr> <chr> <int>
1 177834 A T Still University of Health Sciences Grand.total.men Health.professions.and.related.clinical.sciences...Degrees.total. 212
2 177834 A T Still University of Health Sciences Grand.total.women Health.professions.and.related.clinical.sciences...Degrees.total. 359
3 177834 A T Still University of Health Sciences White.non.Hispanic.men Health.professions.and.related.clinical.sciences...Degrees.total. 181
4 177834 A T Still University of Health Sciences White.non.Hispanic.women Health.professions.and.related.clinical.sciences...Degrees.total. 317
5 177834 A T Still University of Health Sciences Black.non.Hispanic.men Health.professions.and.related.clinical.sciences...Degrees.total. 3
6 177834 A T Still University of Health Sciences Black.non.Hispanic.women Health.professions.and.related.clinical.sciences...Degrees.total. 5
CodePudding user response:
- Using
extractfromtidyrpackage (it is intidyverse) - Capture 2 groups with
() - Define second group to have one or more characters that are not
.up to the end$
library(dplyr)
library(tidyr)
df %>%
extract(CatSex, c("Cat", "Sex"), "(.*)\\.([^.] )$")
UnitID Institution.Name Cat Sex
1 222178 Abilene Christian University Hispanic men
2 222178 Abilene Christian University Hispanic women
3 222178 Abilene Christian University American.Indian.or.Alaska.Native men
4 222178 Abilene Christian University American.Indian.or.Alaska.Native women
5 222178 Abilene Christian University Asian.or.Pacific.Islander women
6 222178 Abilene Christian University Asian.or.Pacific.Islander men
7 222178 Abilene Christian University Grand.total men
8 222178 Abilene Christian University Grand.total women
9 222178 Abilene Christian University White.non.Hispanic men
10 222178 Abilene Christian University White.non.Hispanic women
11 222178 Abilene Christian University lack.non.Hispanic men
12 222178 Abilene Christian University Black.non.Hispanic women
13 222178 Abilene Christian University Hispanic men
14 222178 Abilene Christian University Hispanic women
15 222178 Abilene Christian University American.Indian.or.Alaska.Native men
Disc
1 Communication journalism..and.related.programs
2 Communication journalism and.related.programs
3 Communication journalism..and.related.programs
4 Communication..journalism..and.related.programs
5 Communication..journalism..and.related.programs
6 Communication .journalism..and.related.program
7 Computer.and.information.sciences.and.support.serv
8 computer.and.information.sciences.and.support.servi
9 Computer.and.information.sciences.and.support.servi
10 Computer.and.information.sciences.and.support.servi
11 Computer.and.information.sciences.and.support.servi
12 Computer.and.information.sciences.and.support.servi.
13 Computer.and.information.sciences.and.support.serv
14 Computer.and.information.sciences.and.support.servi.
15 Computer.and.information.sciences.and.support.servi
CodePudding user response:
pivot_longer is not the right function in this context.
Here are few options -
- Using
tidyr::separate
tidyr::separate(df, 'CatSex', c('Cat', 'Sex'), sep = '(\\.)(?!.*\\.)')
#. Cat Sex
#1 Grand.total men
#2 Grand.total women
#3 White.non.Hispanic men
#4 White.non.Hispanic women
#5 Black.non.Hispanic men
#6 Black.non.Hispanic women
- Using
stringrfunctions
library(dplyr)
library(stringr)
df %>%
mutate(Sex = str_extract(CatSex, 'men|women'),
Cat = str_remove(CatSex, '\\.(men|women)'))
- In base R
transform(df, Sex = sub('.*\\.(men|women)', '\\1', CatSex),
Cat = sub('\\.(men|women)', '', CatSex))
data
It is easier to help if you provide data in a reproducible format
df <- data.frame(CatSex = c("Grand.total.men", "Grand.total.women",
"White.non.Hispanic.men", "White.non.Hispanic.women",
"Black.non.Hispanic.men", "Black.non.Hispanic.women"))