Probably easier to understand with an example so here we go:
random = np.random.uniform(size=(3))
what_i_have = pd.DataFrame({
('a', 'a'): random,
('b', 'b1'): np.linspace(3, 5, 3),
('b', 'b2'): np.linspace(6, 8, 3),
('b', 'b3'): np.linspace(9, 11, 3)
})
what_i_want = pd.DataFrame({
('a', 'a'): np.concatenate((random, random, random)),
('b', 'b_category'): ['b1']*3 ['b2']*3 ['b3']*3,
('b', 'b_value'): np.linspace(3, 11, 9)
})
print(what_i_have)
print('----------------------------------')
print(what_i_want)
Output:
a b
a b1 b2 b3
0 0.587075 3.0 6.0 9.0
1 0.798710 4.0 7.0 10.0
2 0.206860 5.0 8.0 11.0
----------------------------------
a b
a b_category b_value
0 0.587075 b1 3.0
1 0.798710 b1 4.0
2 0.206860 b1 5.0
3 0.587075 b2 6.0
4 0.798710 b2 7.0
5 0.206860 b2 8.0
6 0.587075 b3 9.0
7 0.798710 b3 10.0
8 0.206860 b3 11.0
My issue is that my data doesn't just have b1 b2 b3, it also has b4, b5, b6... All the way to about b90. The obvious solution would be to make a loop creating 90 dataframes, one for each category, then concatenating them into one dataframe, but I imagine there must be a better way of doing it.
edit:
what_i_have.unstack() doesn't really solve the issue, as can be seen below. It could be an intermediate step but there's still some work to do with this result before reaching what I want and I don't see much of an advantage in doing this over the loop solution I've previously mentioned:
a a 0 0.587075
1 0.798710
2 0.206860
b b1 0 3.000000
1 4.000000
2 5.000000
b2 0 6.000000
1 7.000000
2 8.000000
b3 0 9.000000
1 10.000000
2 11.000000
CodePudding user response:
Keeping the MultiIndex, might be a better way out there though:
df = df.melt(id_vars=[df.columns[0]], var_name=['b','b_category'], value_name='b_value')
a = df[[('a','a')]]
b = df[['b', 'b_category', 'b_value']].pivot(columns='b').swaplevel(0,1, axis=1)
df = pd.concat([a, b], axis=1)
df.columns = pd.MultiIndex.from_tuples(df.columns)
print(df)
Output:
a b
a b_category b_value
0 0.737076 b1 3.0
1 0.718409 b1 4.0
2 0.269516 b1 5.0
3 0.737076 b2 6.0
4 0.718409 b2 7.0
5 0.269516 b2 8.0
6 0.737076 b3 9.0
7 0.718409 b3 10.0
8 0.269516 b3 11.0
CodePudding user response:
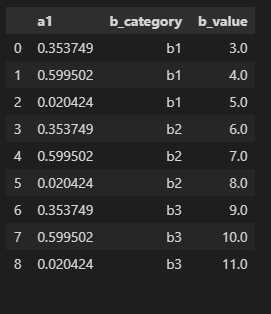
what_i_have.columns = what_i_have.columns.droplevel()
what_i_have.melt(id_vars=['a1'],value_vars=['b1','b2','b3'],var_name='b_category', value_name='b_value')
I think these commands should solve your issue. You lose the multilevel due to the use of the melt. Not sure how to get around that at the moment but otherwise this should meet your requirements.
output
CodePudding user response:
Pull out b and melt:
out = what_i_have.pop('b')
# we need this length to extend
# what_i_have
size = out.columns.size
out = out.melt()
out.columns = ['b_category', 'b_value']
out.columns = [['b', 'b'], out.columns]
Reindex what is left of what_i_have and concatenate:
index = np.tile(what_i_have.a.index, size)
what_i_want = what_i_have.reindex(index).reset_index(drop = True)
pd.concat([what_i_want, out], axis = 1)
a b
a b_category b_value
0 0.883754 b1 3.0
1 0.172427 b1 4.0
2 0.631352 b1 5.0
3 0.883754 b2 6.0
4 0.172427 b2 7.0
5 0.631352 b2 8.0
6 0.883754 b3 9.0
7 0.172427 b3 10.0
8 0.631352 b3 11.0