This is my code:
p14 <- ggplot(plot14, aes(x = Harvest, y = Percentage, fill = factor(Plant, level = orderplants)))
geom_col(show.legend = FALSE)
geom_vline(xintercept=3.5)
labs(y = "Bedekking %",
x = NULL,
fill = "Plantensoort")
theme_classic()
The code is about plant coverage of a plot (I have 70 plots in total). So bedekking is the Dutch word for coverage. The problem: the numbers represent the time periods of the measurements:
- July 2020
- August 2020
- October 2020
- May 2021
- June 2021
- July 2021
- August 2021
- October 2021
I would like the bars of each month to line up, so there would be two rows (2020 and 2021) where the bars of the same months are above/below each other (see ugly sketch below). Is this possible to code, or do I need to change my entire dataset?
CodePudding user response:
It would be better if you could include some sample raw data as part of a reproducible example. I've created a little made up data to illustrate.
Ideally, you'd want the raw dates behind the numbered time measuements so that you can split the dates into months and years as separate variables. Assuming you only have the numbers, you could create some logic like this to make the months and years.
And you could use facet_wrap to show one year above another for the respective months.
library(tidyverse)
library(scales)
tibble(
harvest = seq(1, 8, 1),
percentage = rep(1, 8),
plant = rep(c("this", "that"), 4)
) |>
mutate(
month = case_when(
harvest %in% c(1, 6) ~ 7,
harvest %in% c(2, 7) ~ 8,
harvest %in% c(3, 8) ~ 10,
harvest %in% c(4) ~ 5,
harvest %in% c(5) ~ 6,
TRUE ~ NA_real_
),
year = case_when(
harvest <= 3 ~ 2020,
TRUE ~ 2021
)
) |>
ggplot(aes(month, percentage, fill = plant))
geom_col(show.legend = FALSE)
labs(
y = "Bedekking %",
x = NULL,
fill = "Plantensoort"
)
facet_wrap(~year, ncol = 1)
scale_y_continuous(labels = label_percent())
theme_classic()
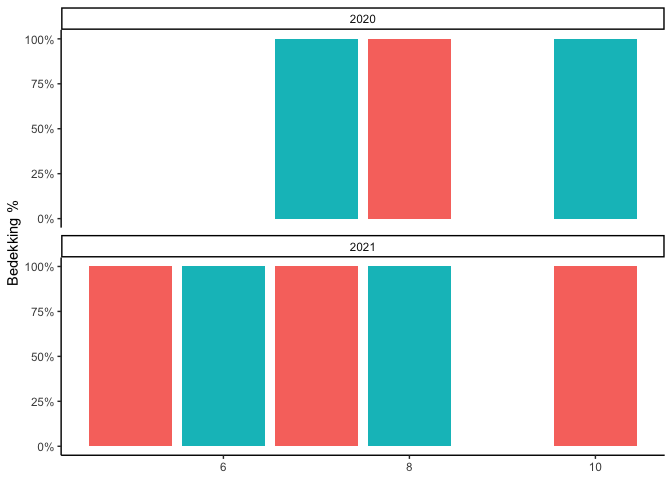
Created on 2022-05-13 by the reprex package (v2.0.1)
