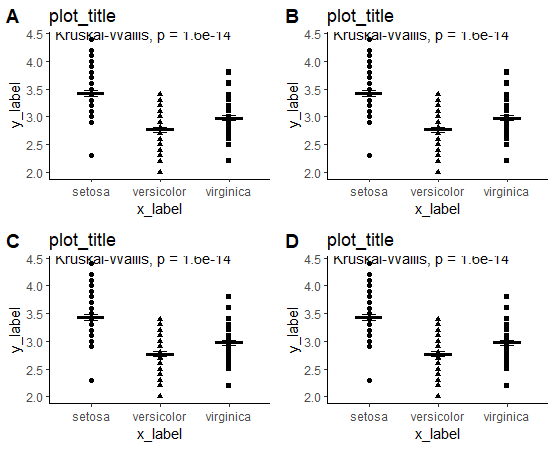
When arranging multiple plots from a plotlist with cowplot, the stat_compare_means values get cut off in the grid (see picture). When I am removing the plots titles it will still be cut off. Is there a way to fix this? Please find reproducible code below:
library(tidyverse)
library(ggplot2)
library(ggpubr)
library(cowplot)
plotlist = list()
u=3
for (i in 0:3) {
single_plot <- iris %>%
ggplot(aes(x = Species, y = Sepal.Width , group=Species)) #create a plot, specify x,y, parameters
geom_point(aes(shape = Species)) # create a
stat_summary(fun = mean, # calculate SEM, specify type and width of the resulting bars
fun.min = function(x) mean(x) - sd(x)/sqrt(length(x)), #calculate ymin SEM
fun.max = function(x) mean(x) sd(x)/sqrt(length(x)), #calculate ymax SEM
geom = 'errorbar', width = 0.2) #specify type of stat_summary and its size
stat_summary(fun = mean, fun.min = mean, fun.max = mean, #calculate mean, specify type, width and size (fatten) of the resulting bars
geom = 'errorbar', width = 0.4, size=1.2) #specify type of stat_summary and its size
labs(x = "x_label", y = "y_label") #set the x- and y-axis labels
ggtitle("plot_title") #set the plot title
theme_classic() #adjust the basic size of the plot
theme(
legend.position = "none", #do not use a plot legend
)
stat_compare_means(method="kruskal.test")
plotlist <- append(plotlist, list(single_plot))
i=i 1
}
plot_grid(plotlist = plotlist,
labels = "AUTO"
)
CodePudding user response:
increase the y axis scale and expand the limits
For this particular plot, I found this combination of parameters to work well.
scale_y_continuous(limits = c(2, 5), expand = c(0, 0.3))
CodePudding user response:
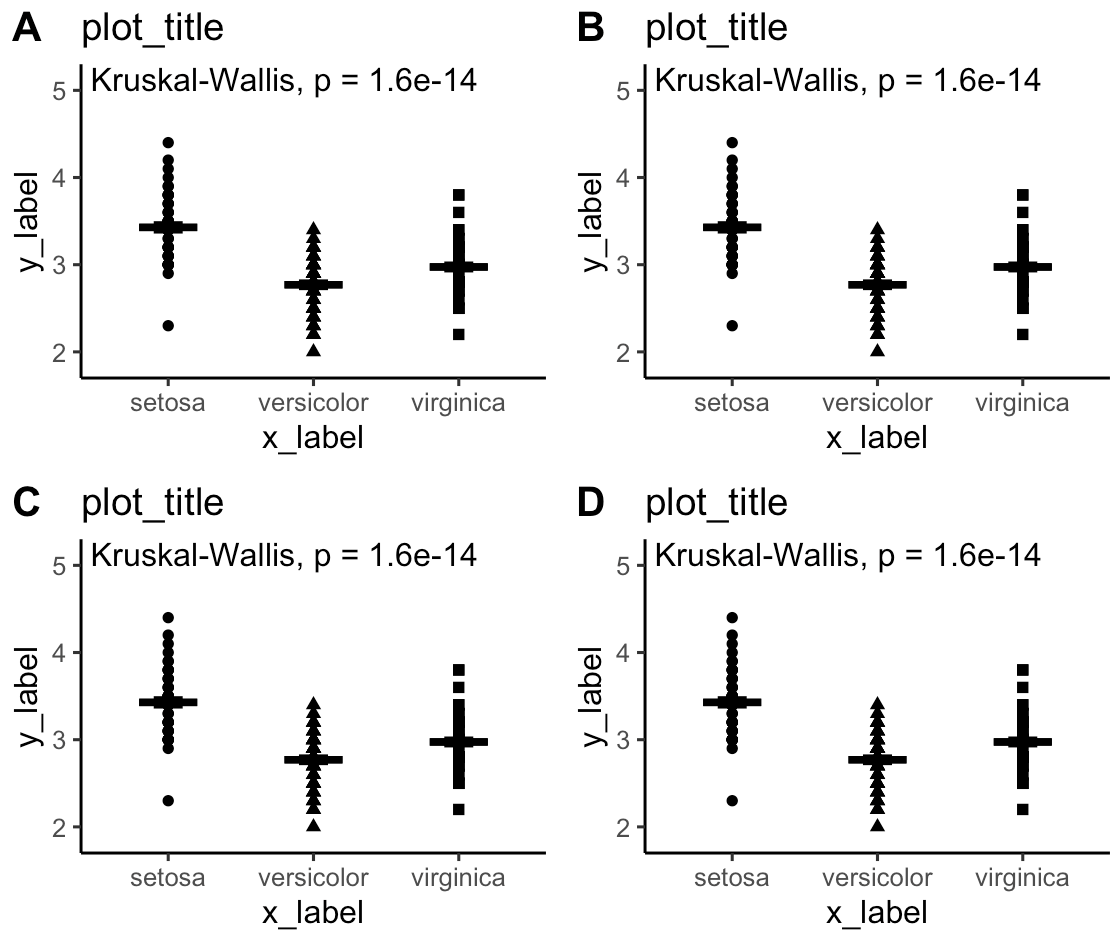
I now figured out the solution. The y-axis has to be scaled using scale_y_continious with a relative expansion using expand(expand = expansion(mult = c(x_expasion, y_expansion). See reproducible code below.
library(tidyverse)
library(ggplot2)
library(ggpubr)
library(cowplot)
plotlist = list()
u=3
for (i in 0:3) {
single_plot <- iris %>%
ggplot(aes(x = Species, y = Sepal.Width , group=Species)) #create a plot, specify x,y, parameters
geom_point(aes(shape = Species)) # create a
stat_summary(fun = mean, # calculate SEM, specify type and width of the resulting bars
fun.min = function(x) mean(x) - sd(x)/sqrt(length(x)), #calculate ymin SEM
fun.max = function(x) mean(x) sd(x)/sqrt(length(x)), #calculate ymax SEM
geom = 'errorbar', width = 0.2) #specify type of stat_summary and its size
stat_summary(fun = mean, fun.min = mean, fun.max = mean, #calculate mean, specify type, width and size (fatten) of the resulting bars
geom = 'errorbar', width = 0.4, size=1.2) #specify type of stat_summary and its size
scale_y_continuous(expand = expansion(mult = c(0,.1))) #create continouos y scale and set relative expansion
labs(x = "x_label", y = "y_label") #set the x- and y-axis labels
ggtitle("plot_title") #set the plot title
theme_classic() #adjust the basic size of the plot
theme(
legend.position = "none", #do not use a plot legend
)
stat_compare_means(method="kruskal.test")
plotlist <- append(plotlist, list(single_plot))
i=i 1
}
print(plot_grid(plotlist = plotlist,
labels = "AUTO"
))