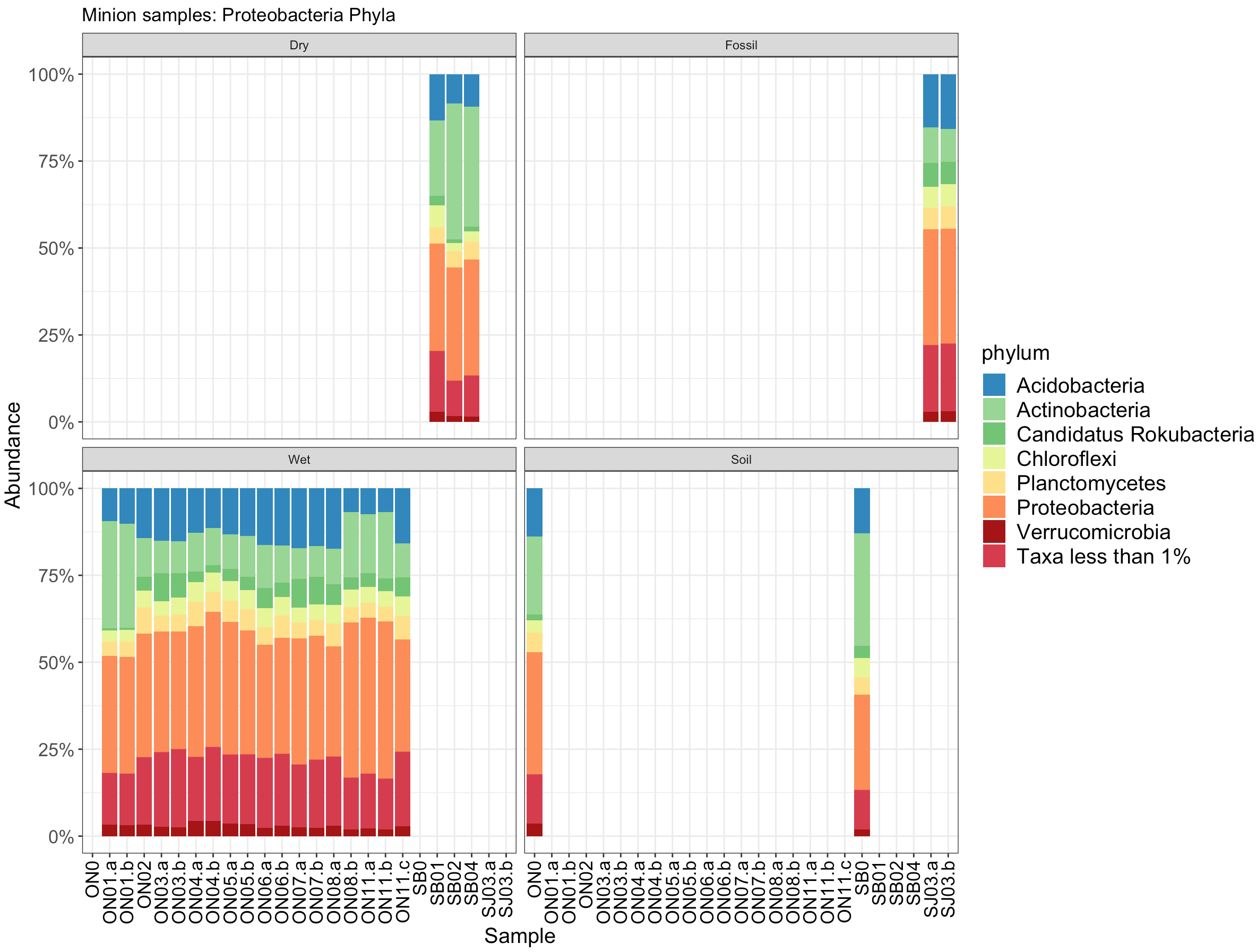
I want to group these bars by the variable "env".
ggplot(dat, aes(x = Sample, fill = phylum, y = Abundance))
geom_bar(position="fill", stat = "identity") theme_bw()
facet_wrap( ~ env)
theme(axis.text.x = element_text(angle = 90, size = 13, colour = "black", vjust = 0.5, hjust = 1), axis.title.x = element_text(size = 15),
axis.text.y = element_text(size = 13, vjust = 0.5, hjust = 1), axis.title.y = element_text(size = 15), legend.title = element_text(size = 15),
legend.text = element_text(size = 15, colour = "black"))
ggtitle("Minion samples: Proteobacteria Phyla") scale_y_continuous(labels = percent_format(), limits=c(0,1))
scale_fill_manual(values = c("Acidobacteria"="#3288bd",
"Actinobacteria" = "#99d594",
"Candidatus Rokubacteria" = "#74c476",
"Chloroflexi"= "#e6f598",
"Planctomycetes"="#fee08b",
"Proteobacteria" = "#fc8d59",
"Verrucomicrobia" = "#a50f15",
"Taxa less than 1%" = "#d53e4f"))
I get this:
I would like to have an unique x-axis and get rid of these empty spaces. Is there an other option beside facet_wrap?
Thanks
CodePudding user response:
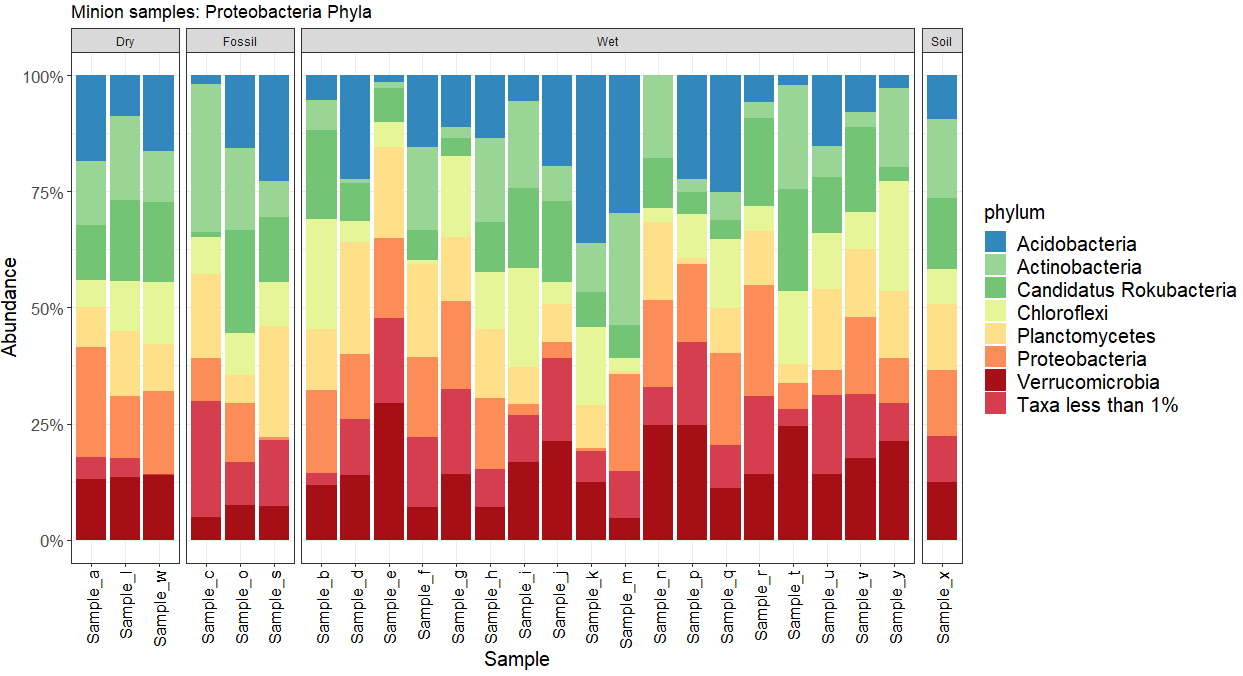
As per the comments, indeed using facet_wrap(~env, scales = "free, nrow =1) will solve your issue.
Moreover, you might want to have an equal width for the histograms, in this case you can use facet_grid and the space argument to have something like this:
ggplot(dat, aes(x = Sample, fill = phylum, y = Abundance))
geom_bar(position="fill", stat = "identity") theme_bw()
facet_grid(. ~ env, scales = "free", space = "free")
theme(axis.text.x = element_text(angle = 90, size = 13, colour = "black", vjust = 0.5, hjust = 1), axis.title.x = element_text(size = 15),
axis.text.y = element_text(size = 13, vjust = 0.5, hjust = 1), axis.title.y = element_text(size = 15), legend.title = element_text(size = 15),
legend.text = element_text(size = 15, colour = "black"))
ggtitle("Minion samples: Proteobacteria Phyla") scale_y_continuous(labels = percent_format(), limits=c(0,1))
scale_fill_manual(values = c("Acidobacteria"="#3288bd",
"Actinobacteria" = "#99d594",
"Candidatus Rokubacteria" = "#74c476",
"Chloroflexi"= "#e6f598",
"Planctomycetes"="#fee08b",
"Proteobacteria" = "#fc8d59",
"Verrucomicrobia" = "#a50f15",
"Taxa less than 1%" = "#d53e4f"))
Whereas, facet_wrap(~ env, scales = "free", nrow = 1) results in unequal width distribution when free on one row:
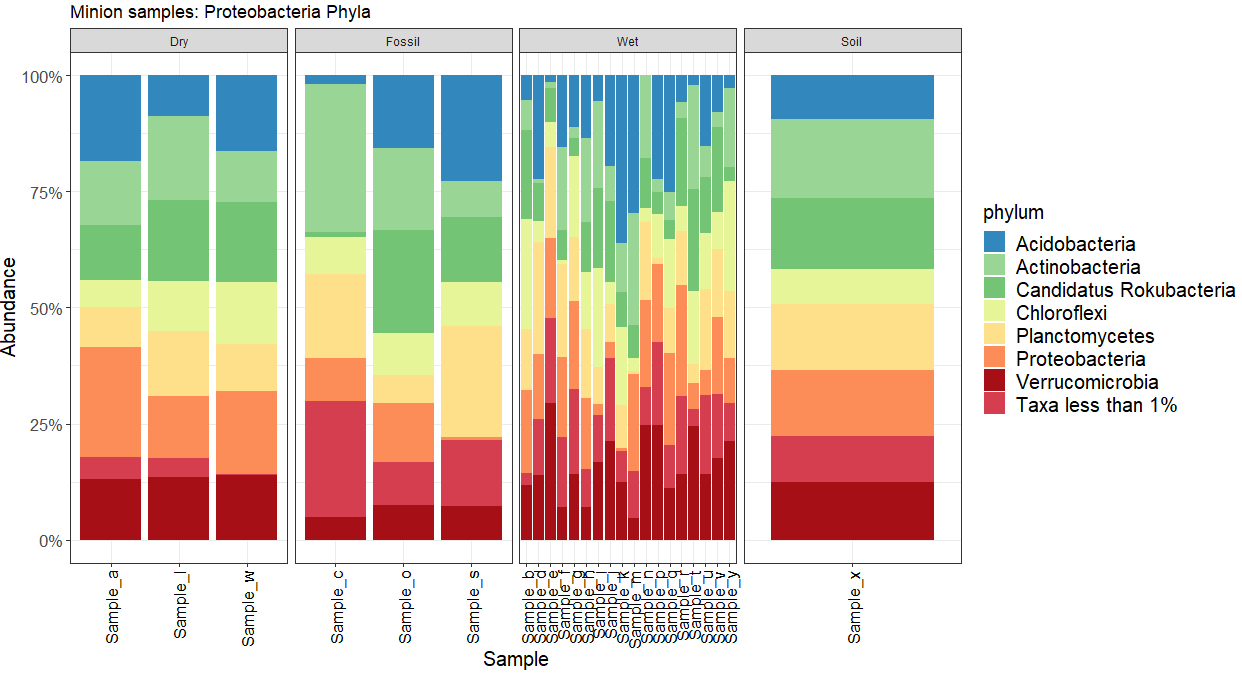
ggplot(dat, aes(x = Sample, fill = phylum, y = Abundance))
geom_bar(position="fill", stat = "identity") theme_bw()
facet_wrap(~ env, scales = "free", nrow = 1)
theme(axis.text.x = element_text(angle = 90, size = 13, colour = "black", vjust = 0.5, hjust = 1), axis.title.x = element_text(size = 15),
axis.text.y = element_text(size = 13, vjust = 0.5, hjust = 1), axis.title.y = element_text(size = 15), legend.title = element_text(size = 15),
legend.text = element_text(size = 15, colour = "black"))
ggtitle("Minion samples: Proteobacteria Phyla") scale_y_continuous(labels = percent_format(), limits=c(0,1))
scale_fill_manual(values = c("Acidobacteria"="#3288bd",
"Actinobacteria" = "#99d594",
"Candidatus Rokubacteria" = "#74c476",
"Chloroflexi"= "#e6f598",
"Planctomycetes"="#fee08b",
"Proteobacteria" = "#fc8d59",
"Verrucomicrobia" = "#a50f15",
"Taxa less than 1%" = "#d53e4f"))
Data
library(scales)
library(ggplot2)
set.seed(18)
dat = data.frame(
env = factor(rep(sample(c("Dry", "Fossil", "Wet", "Soil"), size = 25, prob = c(3,2,18,2), replace = T), each=8), levels =c("Dry", "Fossil", "Wet", "Soil")),
Sample = rep(paste0("Sample_",letters[1:25]), each=8),
phylum = rep(c("Acidobacteria", "Actinobacteria","Candidatus Rokubacteria","Chloroflexi","Planctomycetes", "Proteobacteria", "Verrucomicrobia","Taxa less than 1%"),times = 25),
Abundance = runif(200,0,1))