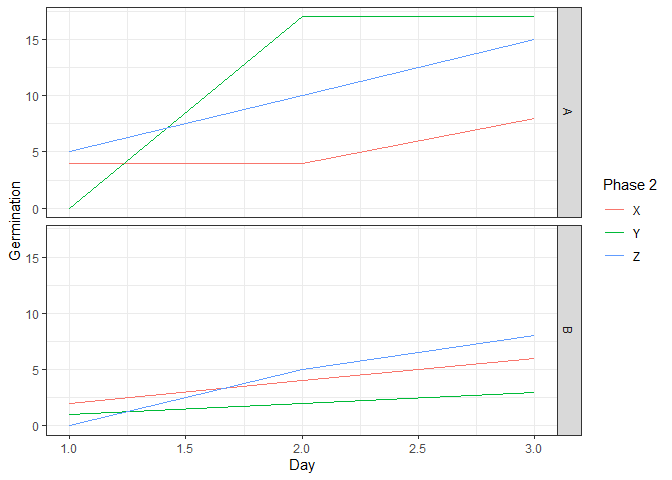
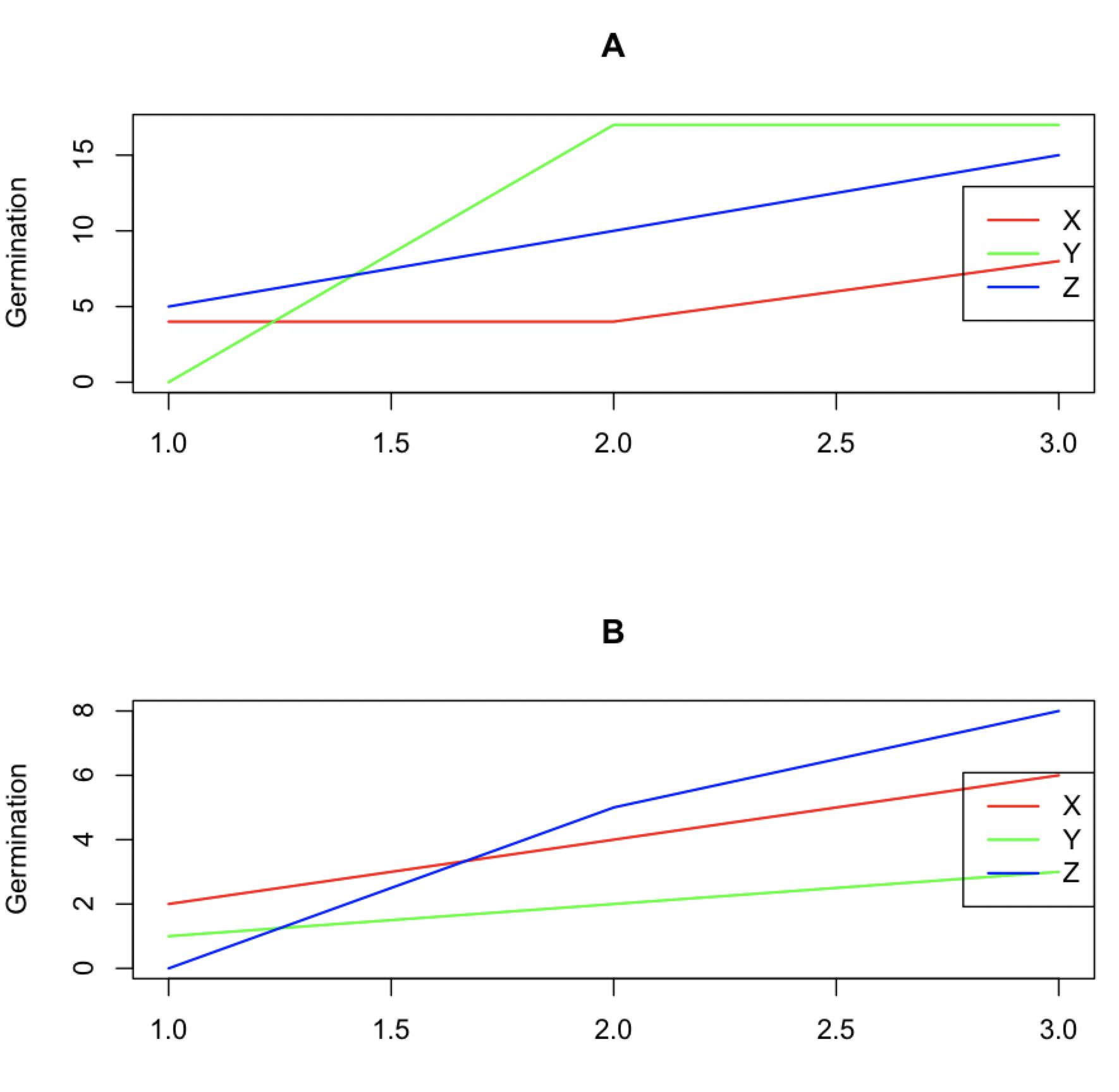
I have data where I am looking at seed germination rates and comparing 2 different treatment phases and seed sources. Each of the 2 phases has multiple groups. I am trying to graph the results as a time series or at least look similar to a time series. The data I have looks like this:
| Source | Phase 1 | Phase 2 | Day 1 | Day 2 | Day 3 |
|---|---|---|---|---|---|
| 1 | A | X | 4 | 4 | 8 |
| 1 | A | Y | 0 | 17 | 17 |
| 1 | A | Z | 5 | 10 | 15 |
| 1 | B | X | 2 | 4 | 6 |
| 1 | B | Y | 1 | 2 | 3 |
| 1 | B | Z | 0 | 5 | 8 |
I have tried converting the data into long form and grouping the day columns using the pivot_longer and names_to functions and then tried graphing using the following code:
ggplot(df1, aes(x= days_num, y = germination, group=phase 1))
geom_line(aes(color=phase 2))
facet_grid(source~.)
It gives me a graph like this: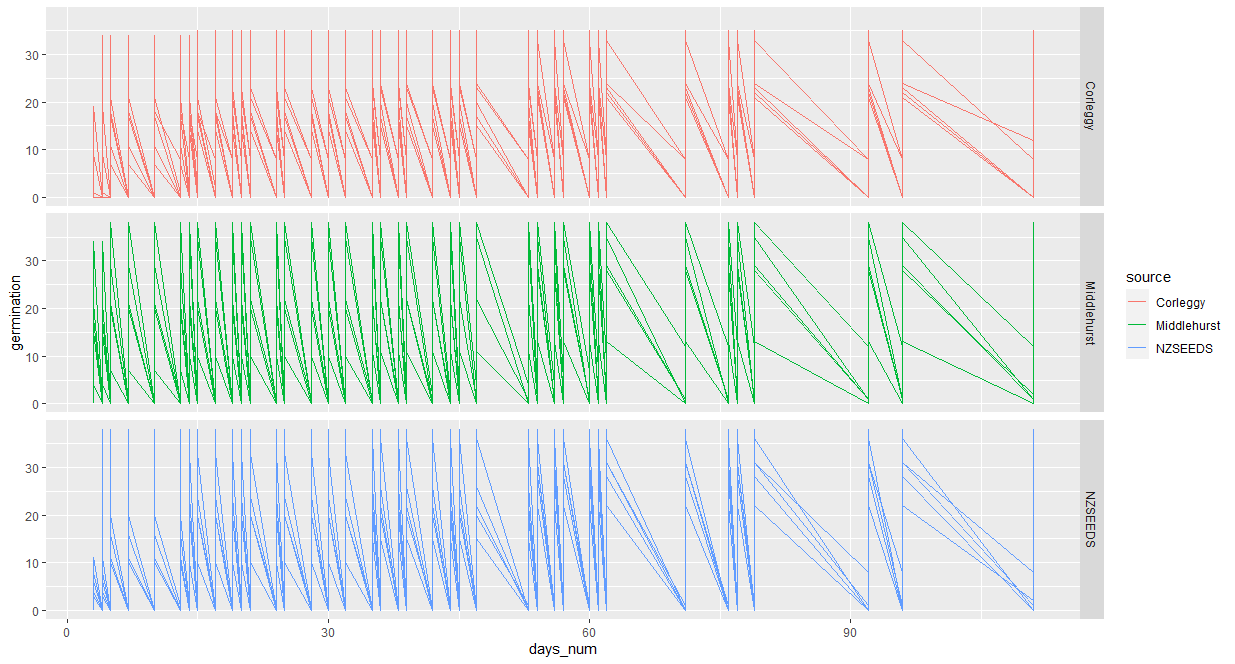
The following image is along the lines of what I would like it to look like: 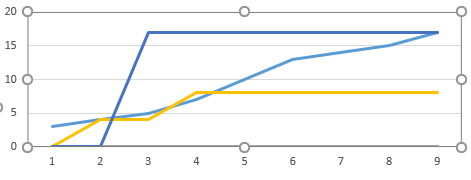
I'd like to be able to have my different sources in graphs side by side, and then have my phase 1 (A - XYZ) lines as one graph and (B - XYZ) separate.
Can anyone help with this?
CodePudding user response:
When mapping phase 1 to group, you are mapping 2 values to each of 3 different phases 2. Map phase 2 to color and facet the plot by phase 1 instead.
df1 <- "Source 'Phase 1' 'Phase 2' 'Day 1' 'Day 2' 'Day 3'
1 A X 4 4 8
1 A Y 0 17 17
1 A Z 5 10 15
1 B X 2 4 6
1 B Y 1 2 3
1 B Z 0 5 8"
df1 <- read.table(text = df1, header = TRUE, check.names = FALSE)
suppressPackageStartupMessages({
library(ggplot2)
library(dplyr)
library(tidyr)
})
df1 %>%
pivot_longer(starts_with("Day"), names_to = "day_num", values_to = "germination") %>%
mutate(day_num = stringr::str_extract(day_num, "\\d "),
day_num = as.integer(day_num)) %>%
ggplot(aes(day_num, germination, color = `Phase 2`))
geom_line()
labs(x = "Day", y = "Germination")
facet_grid(rows = vars(`Phase 1`))
theme_bw()