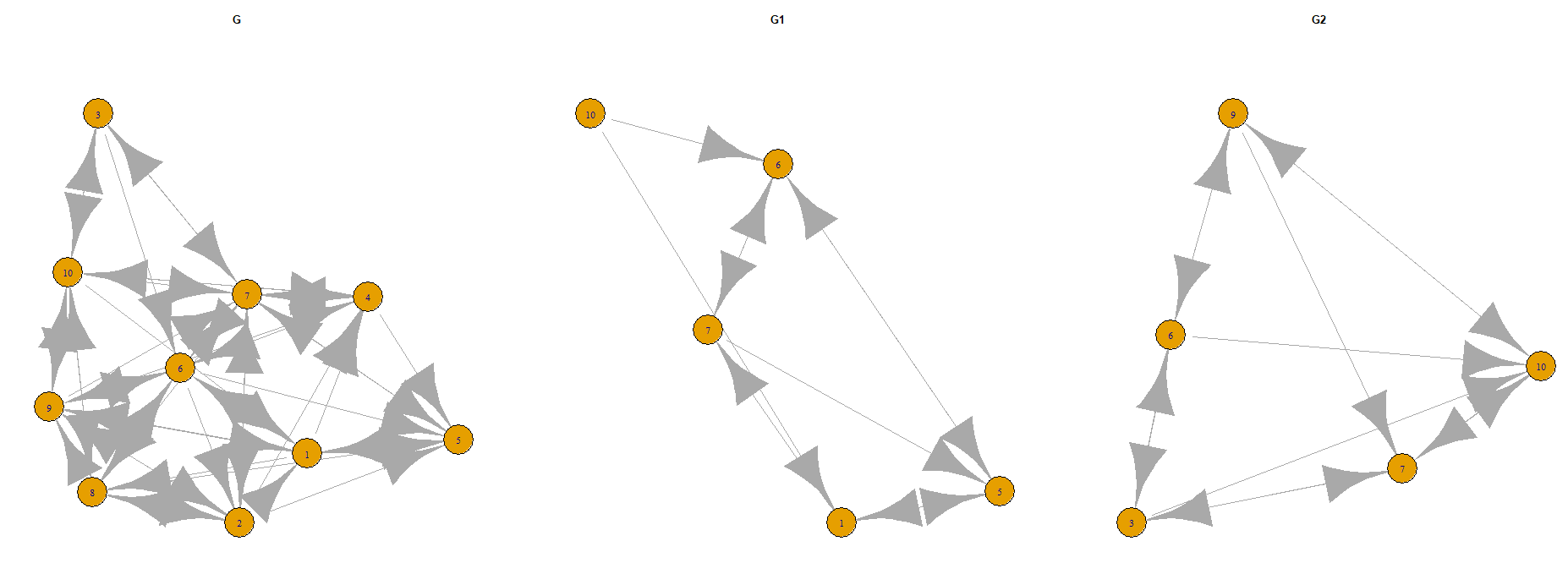
I have a igraph G, I need to sample two overlapping subgraphs G1, G2 and to plot them on the graph with the same layout.
My attempt is below:
library(igraph)
set.seed(1)
n <- 10
A <- matrix(sample(0:1, n * n, rep=TRUE), n, n)
diag(A) = 0
g <- graph_from_adjacency_matrix(A)
V(g)$names <- c(1:n)
id1 = sample(V(g), size = n %/% 2, replace = FALSE)
id2 = sample(V(g), size = n %/% 2, replace = FALSE)
g1 <- induced_subgraph(g, vids = id1)
g2 <- induced_subgraph(g, vids = id2)
V(g1)$names <- c(id1)
V(g2)$names <- c(id2)
#V(g)[id1]$color <- "red"
#V(g)[id2]$color <- "green"
par(mfrow=c(1,3))
layout <- layout.fruchterman.reingold(g)
plot(g, layout=layout, main="G")
plot(g1, layout = layout[-c(setdiff(1:n, id1)),], vertex.label=V(g)[id1], main="G1")
plot(g2, layout = layout[-c(setdiff(1:n, id2)),], vertex.label=V(g)[id2], main="G2")
My problem with ids and labels.
Question. How to plot the igraph subgraphs with saving the nodes' positions and ids?
CodePudding user response:
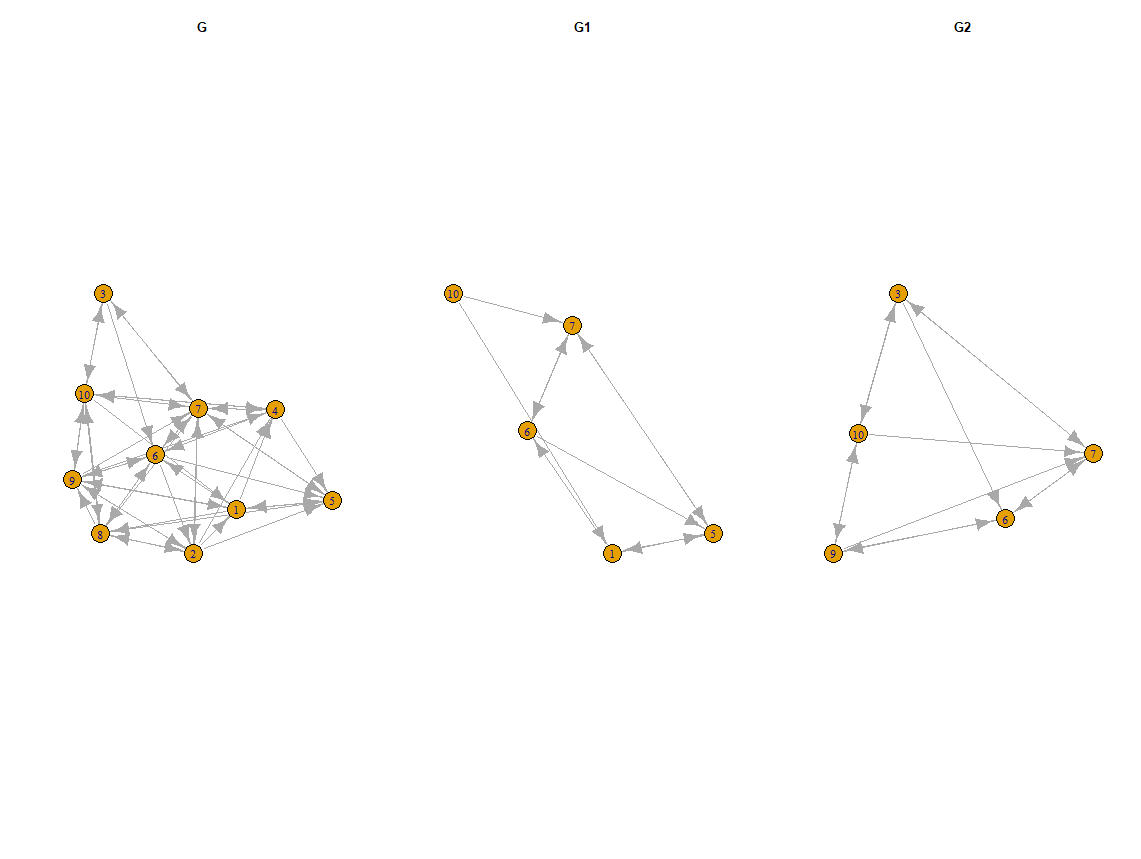
Try the code below
library(igraph)
set.seed(1)
n <- 10
A <- matrix(sample(0:1, n * n, rep = TRUE), n, n)
diag(A) <- 0
g <- graph_from_adjacency_matrix(A)
id1 <- sort(as.integer(sample(V(g), size = n %/% 2, replace = FALSE)))
id2 <- sort(as.integer(sample(V(g), size = n %/% 2, replace = FALSE)))
g1 <- induced_subgraph(g, vids = id1)
g2 <- induced_subgraph(g, vids = id2)
par(mfrow = c(1,3))
layout <- layout.fruchterman.reingold(g)
layout2 <- layout[id2, ]
plot(g, layout = layout, main = "G")
plot(g1, layout = layout[id1, ], main = "G1")
plot(g2, layout = layout[id2, ], main = "G2")