Here is a DNA string that I want to split and then combine in groups of 3
dna=c("TACACGATGACAGTCTTGACGGGTTCTCCTACT")
dna.sg = unlist(strsplit(dna, ""))
Gives
[1] "T" "A" "C" "A" "C" "G" "A" "T" "G" "A" "C" "A" "G" "T" "C" "T" "T" "G" "A" "C" "G" "G" "G" "T" "T" "C" "T" "C" "C" "T" "A" "C" "T"
But I'd like to have
"TAC" "ACG" [...]
CodePudding user response:
You may split every 3 characters in strsplit.
unlist(strsplit(dna, "(?<=.{3})", perl = TRUE))
#[1] "TAC" "ACG" "ATG" "ACA" "GTC" "TTG" "ACG" "GGT" "TCT" "CCT" "ACT"
CodePudding user response:
Other possibilities:
dna <- c("TACACGATGACAGTCTTGACGGGTTCTCCTACT")
regmatches(dna, gregexpr(".{3}", dna))[[1]]
# [1] "TAC" "ACG" "ATG" "ACA" "GTC" "TTG" "ACG" "GGT" "TCT" "CCT" "ACT"
sapply(seq(1, nchar(dna), 3), \(x) substr(dna, x, x 3-1))
# [1] "TAC" "ACG" "ATG" "ACA" "GTC" "TTG" "ACG" "GGT" "TCT" "CCT" "ACT"
substring(dna, seq(1, nchar(dna), by = 3), seq(3, nchar(dna), by = 3))
# [1] "TAC" "ACG" "ATG" "ACA" "GTC" "TTG" "ACG" "GGT" "TCT" "CCT" "ACT"
unlist(strsplit(gsub("(.{3})", "\\1 ", dna), split = " "))
# [1] "TAC" "ACG" "ATG" "ACA" "GTC" "TTG" "ACG" "GGT" "TCT" "CCT" "ACT"
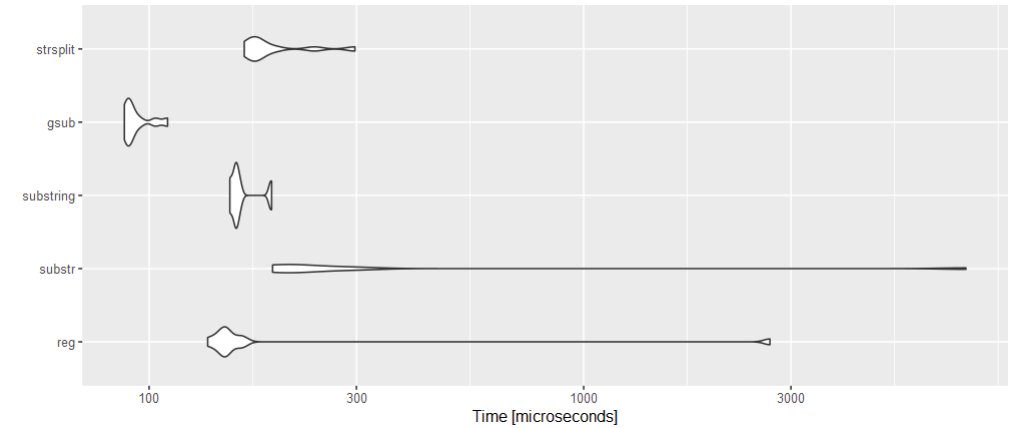
Felt like doing a benchmarking with all those solutions:
dna <- c("TACACGATGACAGTCTTGACGGGTTCTCCTACT")
library(microbenchmark)
bm <- microbenchmark(
reg = regmatches(dna, gregexpr(".{3}", dna))[[1]],
substr = sapply(seq(1, nchar(dna), 3), \(x) substr(dna, x, x 3-1)),
substring = substring(dna, seq(1, nchar(dna), by = 3), seq(3, nchar(dna), by = 3)),
gsub = unlist(strsplit(gsub("(.{3})", "\\1 ", dna), split = " ")),
strsplit = unlist(strsplit(dna, "(?<=.{3})", perl = TRUE)),
times = 10L,
setup = gc(FALSE)
)
autoplot(bm)
gsub seems to be a clear winner!