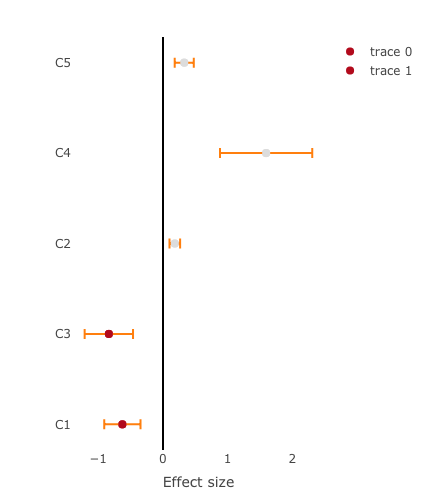
I have a data.frame of regression coefficients with the associated p-values:
library(dplyr)
set.seed(1)
effects.df <- data.frame(contrast = paste0("C",1:5), effect = rnorm(5), stringsAsFactors = F) %>%
dplyr::mutate(effect.error = abs(effect)/sqrt(5)) %>%
dplyr::mutate(p.value = pnorm(effect/effect.error)) %>%
dplyr::arrange(p.value)
effects.df$contrast <- factor(effects.df$contrast,levels = effects.df$contrast)
Which I want to display as a forest plot (X-axis are the effect size and Y-axis are the 'contrast's), where the points and their associated error bars (effect.error) are color coded by 1-p.value, using R's plotly.
Here's what I'm trying:
library(plotly)
effects.plot <- plot_ly(x = effects.df$effect, y = effects.df$contrast, type = 'scatter', mode = "markers", marker = list(size = 8, colorbar = "Hot", color = 1-effects.df$p.value)) %>%
layout(xaxis=list(title = "Effect size",zerolinewidth = 2, zerolinecolor = plotly::toRGB('black'), showgrid = F), yaxis = list(showgrid = F)) %>%
add_trace(error_x = list(array = effects.df$effect.error, width = 5),marker = list(size = 8,colorbar = "Hot", color = 1-effects.df$p.value))
It's close because it's color-coding the points how I want them to but not the error bars.
Any idea how to:
- Color the error bars similar to the points?
- Get the color-bar to show?
CodePudding user response:
I'm not sure that it will allow you to color the error bars separately without some (a lot) of creativity. If you created separate traces for each color, you might be able to force it to comply.
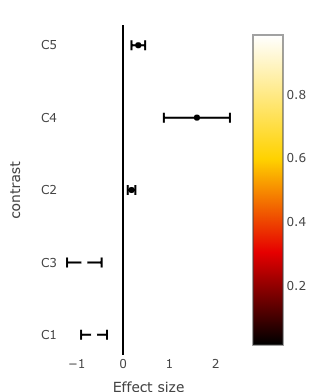
There are many ways you could show the color bar. Here's one way:
(effects.plot <- plot_ly(data = effects.df,
x = ~effect,
y = ~contrast,
error_x = list(array = ~effect.error,
width = 5,
color = "black"),
type = 'scatter',
mode = "markers",
marker = list(colorscale = "Hot",
colorbar = list(size = 8),
color = 1 - effects.df$p.value)) %>%
layout(xaxis=list(title = "Effect size",
zerolinewidth = 2,
zerolinecolor = plotly::toRGB('black'),
showgrid = F),
yaxis = list(showgrid = F)) # set the joined color axis
)
By the way, I noticed that the colors you have are gray and red, not black and white, as shown in my image. You're getting a different color scale than you were expecting.
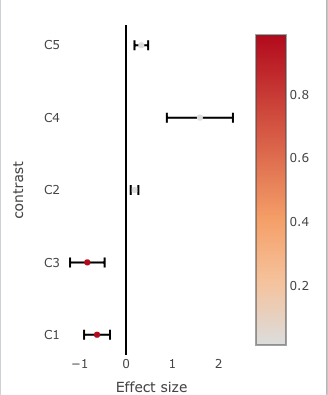
You can see what I mean by plotting this a different way:
(effects.plot <- plot_ly(data = effects.df,
x = ~effect,
y = ~contrast,
error_x = list(array = ~effect.error,
width = 5,
color = "black"),
type = 'scatter',
mode = "markers",
marker = list(coloraxis = "coloraxis",
color = 1 - effects.df$p.value)) %>%
layout(xaxis=list(title = "Effect size",
zerolinewidth = 2,
zerolinecolor = plotly::toRGB('black'),
showgrid = F),
yaxis = list(showgrid = F),
coloraxis = list(colorbar = "Hot", size = 8))
)
This plot is not using the "Hot" color scale. That scale is shown in the first image.
CodePudding user response:
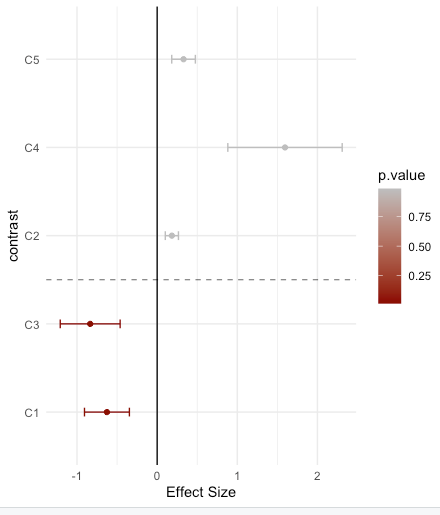
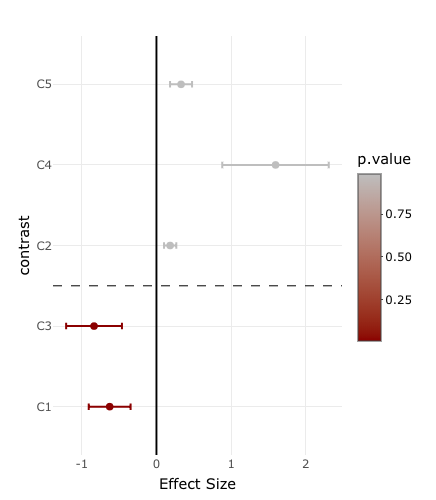
The easiest way to solve this is to use ggplot2 and then to convert it to a plotly object:
Libraries and data:
library(dplyr)
library(plotly)
library(ggplot2)
set.seed(1)
effects.df <- data.frame(contrast = paste0("C",1:5), effect = rnorm(5), stringsAsFactors = F) %>%
dplyr::mutate(effect.error = abs(effect)/sqrt(5)) %>%
dplyr::mutate(p.value = pnorm(effect/effect.error)) %>%
dplyr::arrange(p.value)
Here I also add a horizontal dashed y-line to mark the p-value = 0.05 cutoff:
effects.df$contrast <- factor(effects.df$contrast,levels=effects.df$contrast)
y.intercept <- min(which(effects.df$p.value > 0.05))-0.5
pp <- ggplot(effects.df) geom_vline(xintercept=0,color="black") geom_point(aes(y=contrast,x=effect,color=p.value))
geom_errorbarh(aes(y=contrast,xmin=effect-effect.error,xmax=effect effect.error,x=effect,color=p.value,height=0.1))
scale_color_continuous(low="darkred",high="gray") theme_minimal() xlab("Effect Size")
geom_hline(yintercept=y.intercept,linetype="dashed",color="black",size=0.25)
And the plotly object:
ggplotly(pp)