My data (short example) looks like this:
dput(head(sdNDVI,16))
structure(list(Date = structure(c(19095, 19095, 19096, 19096,
19097, 19097, 19098, 19098, 19099, 19099, 19100, 19100, 19101,
19101, 19102, 19102), class = "Date"), target = c("grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree"), d_sd = c(0.009830432,
0.00614358, 0.001431813, 0.003589623, 0.001397262, 0.001755547,
0.001484436, 0.001647977, 0.002919854, 0.004303091, 0.002274095,
0.005073607, 0.002392742, 0.010799317, 0.002125828, 0.015356333
), NL_sd = c(0.011630472, 0.006169669, 0.001409577, 0.003623346,
0.001371575, 0.001770516, 0.001443259, 0.001662584, 0.002886186,
0.00429965, 0.002226, 0.005068018, 0.002423769, 0.010747391,
0.001948935, 0.014982166), NLncor_sd = c(0.009883439, 0.006186777,
0.001439041, 0.003614421, 0.001404336, 0.001767561, 0.001491952,
0.001658882, 0.002934716, 0.004330486, 0.002285389, 0.005105774,
0.002404389, 0.010864433, 0.00213608, 0.015449409), min_sd = c(0.009830432,
0.00614358, 0.001409577, 0.003589623, 0.001371575, 0.001755547,
0.001443259, 0.001647977, 0.002886186, 0.00429965, 0.002226,
0.005068018, 0.002392742, 0.010747391, 0.001948935, 0.014982166
)), row.names = c(NA, 16L), class = "data.frame")
The column min_sd represents the minimum value per row of the 3 columns d_sd, NL_sd, NLnocor_sd.
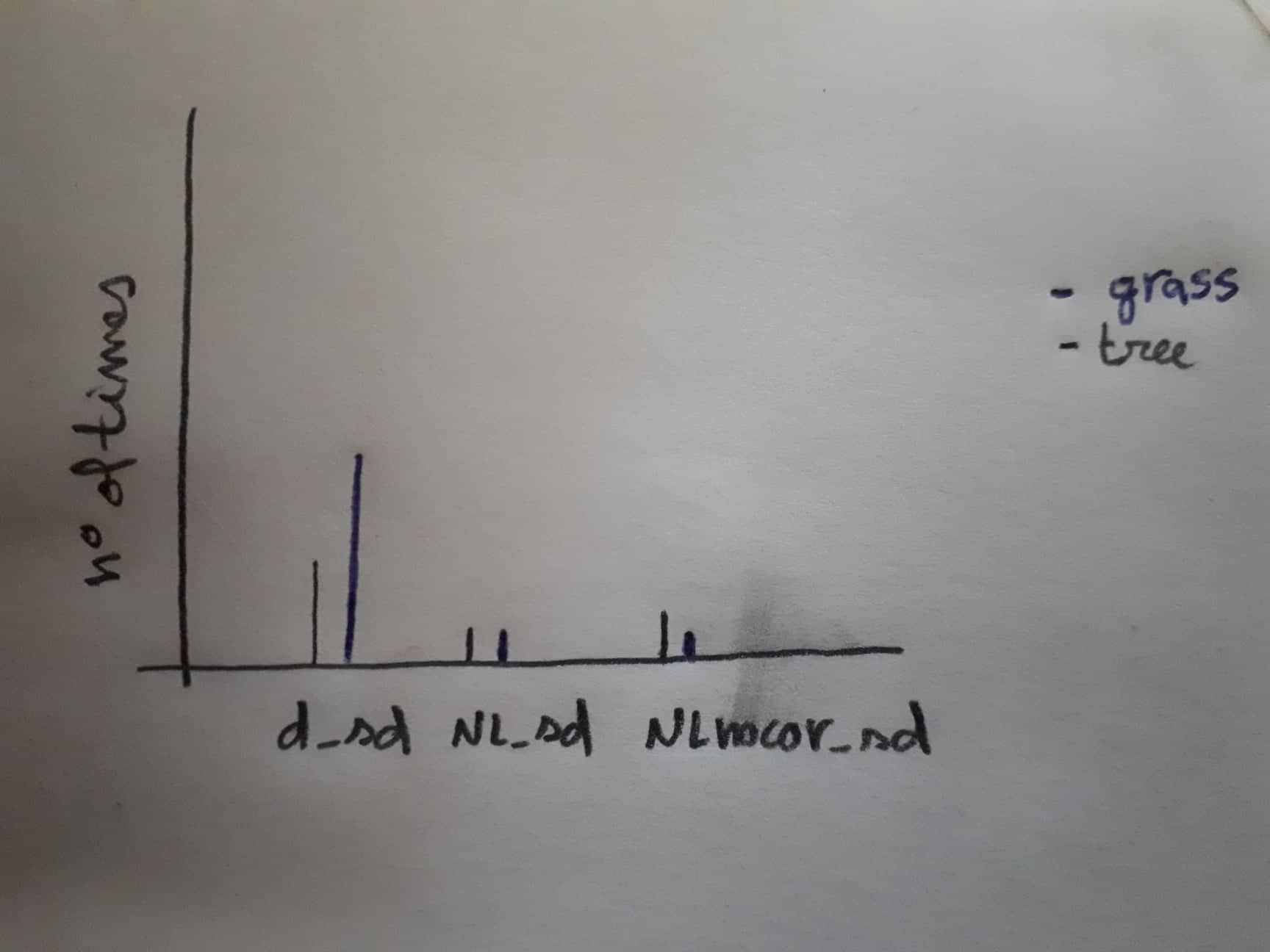
Now I wanted to do a barplot (horizontal or vertical) to represent the number of times each column of the 3 d_sd, NL_sd, NLnocor_sd are represented in min_sd. Each one of the 3 bar plot should be sub-divided by target, grass and tree.
The desired plot output would be something like this (sorry for the drawing):
I'm used to working with ggplot2 but if you use other tools I will be receptive as well. Thank you in advance.
After trying:
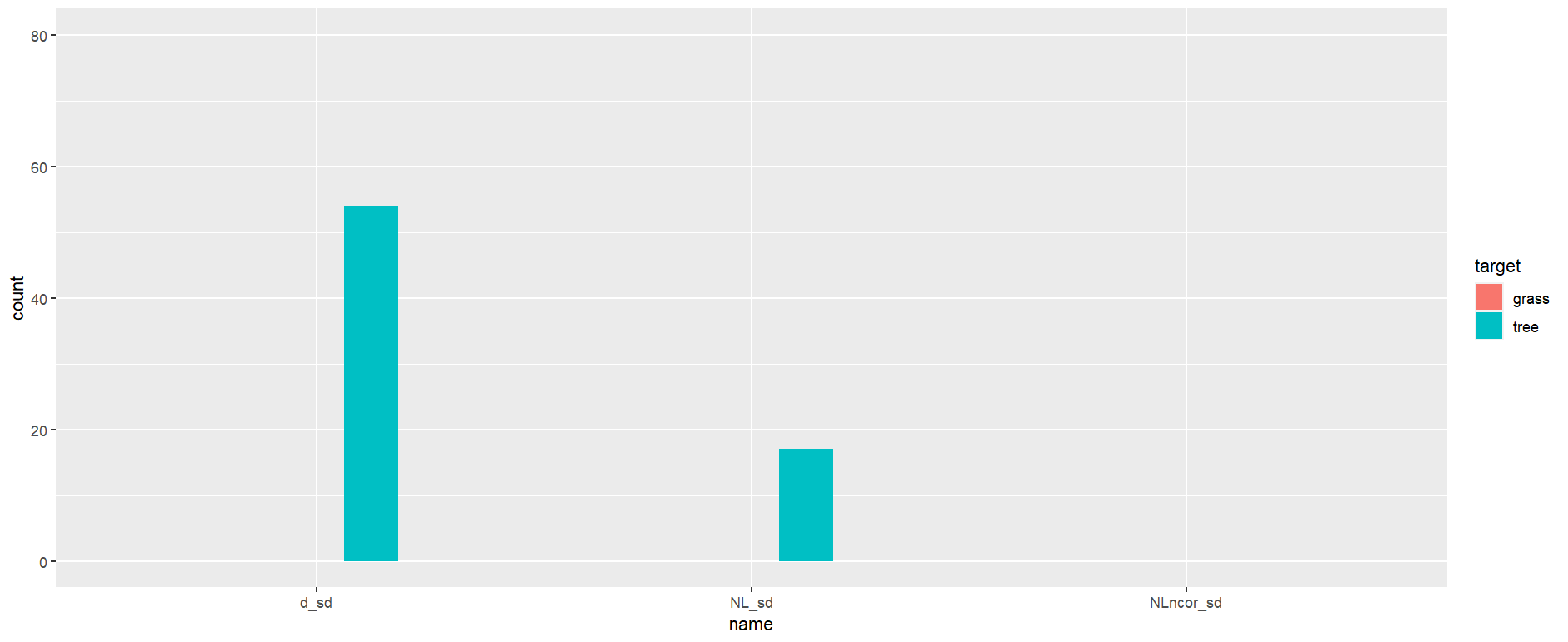
sdNDVI %>% mutate(across(3:5, ~ifelse(.x == min_sd, 1, 0))) %>% pivot_longer(c(3:5)) %>% group_by(target, name) %>% summarise(count = sum(value)) %>% ggplot(aes(x = name, fill = target, y = count)) geom_col(width = 0.25, position = position_dodge(width = 0.5), na.rm = TRUE) scale_y_continuous(limits = c(0, 80))
dput(head(sdNDVI, 142))
structure(list(Date = structure(c(19095, 19095, 19096, 19096,
19097, 19097, 19098, 19098, 19099, 19099, 19100, 19100, 19101,
19101, 19102, 19102, 19103, 19103, 19104, 19104, 19105, 19105,
19106, 19106, 19107, 19107, 19108, 19108, 19109, 19109, 19110,
19110, 19111, 19111, 19112, 19112, 19113, 19113, 19114, 19114,
19115, 19115, 19116, 19116, 19117, 19117, 19118, 19118, 19119,
19119, 19120, 19120, 19121, 19121, 19122, 19122, 19123, 19123,
19124, 19124, 19125, 19125, 19126, 19126, 19127, 19127, 19128,
19128, 19129, 19129, 19130, 19130, 19131, 19131, 19132, 19132,
19133, 19133, 19134, 19134, 19135, 19135, 19136, 19136, 19137,
19137, 19138, 19138, 19139, 19139, 19140, 19140, 19141, 19141,
19142, 19142, 19143, 19143, 19144, 19144, 19145, 19145, 19146,
19146, 19147, 19147, 19148, 19148, 19149, 19149, 19150, 19150,
19151, 19151, 19152, 19152, 19153, 19153, 19154, 19154, 19155,
19155, 19156, 19156, 19157, 19157, 19158, 19158, 19159, 19159,
19160, 19160, 19161, 19161, 19162, 19162, 19163, 19163, 19164,
19164, 19165, 19165), class = "Date"), target = c("grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree", "grass", "tree", "grass", "tree",
"grass", "tree", "grass", "tree"), d_sd = c(0.009830432, 0.00614358,
0.001431813, 0.003589623, 0.001397262, 0.001755547, 0.001484436,
0.001647977, 0.002919854, 0.004303091, 0.002274095, 0.005073607,
0.002392742, 0.010799317, 0.002125828, 0.015356333, 0.00489519,
0.005432558, 0.031744356, 0.013330765, 0.004380233, 0.008529259,
0.009236926, 0.002794691, 0.004381456, 0.005029592, 0.022307418,
0.017924634, 0.015184163, 0.012527571, 0.02126106, 0.003522298,
0.004182938, 0.002314368, 0.0076246, 0.002879415, 0.004333378,
0.003153228, 0.028533612, 0.014873218, 0.013453042, 0.00509009,
0.011874007, 0.00307663, 0.008828247, 0.005550763, 0.005968105,
0.008279004, 0.005995636, 0.008729083, 0.005397178, 0.009258435,
0.005674674, 0.00849763, 0.005765362, 0.014347054, 0.005194654,
0.010036212, 0.005899384, 0.00962091, 0.006742776, 0.010771743,
0.006208567, 0.027574999, 0.016282077, 0.015474263, 0.014341471,
0.016104659, 0.009962326, 0.018895618, 0.007288482, 0.011912717,
0.004340415, 0.003491596, 0.005178044, 0.00164198, 0.010895453,
0.014226956, 0.007875839, 0.002976035, 0.005950366, 0.019527251,
0.007573006, 0.039021655, 0.00434444, 0.008165457, 0.002771065,
0.002549085, NA, 0.00068218, 0.002355381, 0.00795954, 0.005759183,
0.005016125, 0.002944426, 0.0084233, 0.006860759, 0.02614528,
0.026728134, 0.018955912, 0.00378909, 0.014559335, 0.004812223,
0.015360884, 0.011666986, 0.009930083, 0.0041961, 0.006446649,
0.00237411, 0.007755967, 0.001878764, 0.010208893, 0.012109833,
0.019613881, 0.002148972, 0.009214464, 0.001980726, 0.005662194,
0.001899928, 0.005803247, 0.002201519, 0.00328676, 0.002014854,
0.004649152, 0.002274161, 0.004801268, 0.00205222, 0.033229958,
0.020512938, 0.005806449, 0.002866929, 0.004925562, 0.002816567,
0.01016808, 0.004163812, 0.007851256, 0.004111843, 0.01091895,
0.002867563, 0.007332856, 0.031817319, 0.017612059), NL_sd = c(0.011630472,
0.006169669, 0.001409577, 0.003623346, 0.001371575, 0.001770516,
0.001443259, 0.001662584, 0.002886186, 0.00429965, 0.002226,
0.005068018, 0.002423769, 0.010747391, 0.001948935, 0.014982166,
0.004788058, 0.005418707, 0.031635842, 0.013425189, 0.004290994,
0.008584524, 0.009329015, 0.00282333, 0.004385241, 0.005214086,
0.02178649, 0.018136813, 0.014964051, 0.012455305, 0.017578538,
0.00353749, 0.003995365, 0.002304488, 0.007702759, 0.002853387,
0.004211933, 0.003113751, 0.028071121, 0.014845754, 0.013223408,
0.004993331, 0.011854977, 0.003165374, 0.009051744, 0.005489159,
0.005821341, 0.008176441, 0.005847643, 0.008632582, 0.005255749,
0.009172938, 0.005514234, 0.008449149, 0.005600061, 0.014342186,
0.005054358, 0.010086637, 0.005694254, 0.009723672, 0.006520761,
0.010932219, 0.005989528, 0.028083292, 0.015720081, 0.016008612,
0.01363116, 0.016686721, 0.009909327, 0.019530434, 0.006892656,
0.012533604, 0.004188733, 0.003737502, 0.004924819, 0.001759126,
0.010121838, 0.015178924, 0.007660564, 0.003224047, 0.005713842,
0.020653574, 0.007317763, 0.040863774, 0.004191221, 0.008730109,
0.002657193, 0.002757261, NA, 0.000737879, 0.002252117, 0.00854629,
0.005629613, 0.005398059, 0.002790605, 0.009009766, 0.006397561,
0.027547179, 0.025029065, 0.020227057, 0.003711371, 0.015552794,
0.004751886, 0.016376306, 0.009555682, 0.010632038, 0.004168767,
0.006922182, 0.002351582, 0.00832403, 0.00180232, 0.010915895,
0.010828096, 0.020690998, 0.002060375, 0.00981494, 0.001909286,
0.006080086, 0.0018259, 0.006178081, 0.002112633, 0.003538175,
0.001935102, 0.004993449, 0.002166196, 0.005150362, 0.001954427,
0.034646982, 0.019154351, 0.006266365, 0.00259939, 0.005227667,
0.002736231, 0.010738438, 0.002615967, 0.008236663, 0.003662972,
0.011503301, 0.002805302, 0.007725738, 0.030372904, 0.018750948
), NLncor_sd = c(0.009883439, 0.006186777, 0.001439041, 0.003614421,
0.001404336, 0.001767561, 0.001491952, 0.001658882, 0.002934716,
0.004330486, 0.002285389, 0.005105774, 0.002404389, 0.010864433,
0.00213608, 0.015449409, 0.00491889, 0.005466893, 0.031936029,
0.013420962, 0.004402015, 0.00858508, 0.009281366, 0.002812714,
0.00440264, 0.005061974, 0.022421449, 0.018043392, 0.01526026,
0.012607695, 0.021343068, 0.003544158, 0.004202317, 0.002328418,
0.007660857, 0.002896304, 0.004353618, 0.003171484, 0.028684822,
0.014964763, 0.013520917, 0.005120415, 0.011933198, 0.003094631,
0.008871585, 0.005582728, 0.005996811, 0.008326673, 0.0060241,
0.008780066, 0.005421961, 0.009313822, 0.005699835, 0.008550701,
0.005789815, 0.014439645, 0.005216259, 0.010104136, 0.005922856,
0.009688467, 0.006769191, 0.010849978, 0.006231775, 0.027778982,
0.016347691, 0.015595033, 0.014398889, 0.016233601, 0.010001014,
0.019049073, 0.007315738, 0.012012295, 0.004355549, 0.003522151,
0.005196072, 0.001656396, 0.010933954, 0.014350064, 0.007903317,
0.003002422, 0.005970026, 0.019693402, 0.007597911, 0.03934434,
0.004358105, 0.008236689, 0.002779326, 0.002571942, NA, 0.000688279,
0.002361943, 0.008029853, 0.005776324, 0.0050605, 0.002952805,
0.008497156, 0.006879978, 0.026365373, 0.026813316, 0.019120611,
0.0037996, 0.014686509, 0.00482537, 0.015494039, 0.011698044,
0.01001808, 0.00420703, 0.006503711, 0.002380033, 0.00782429,
0.001883311, 0.010298417, 0.01213804, 0.019779949, 0.002154125,
0.009294659, 0.001985369, 0.005712263, 0.00190437, 0.00585404,
0.002206569, 0.00331604, 0.002019494, 0.00469033, 0.002279385,
0.004843563, 0.002057064, 0.033500339, 0.02057835, 0.00585848,
0.002873707, 0.004968185, 0.002822988, 0.010255357, 0.00417444,
0.007916964, 0.004121695, 0.011009909, 0.002874513, 0.007393927,
0.031909371, 0.01776547), min_sd = c(0.009830432, 0.00614358,
0.001409577, 0.003589623, 0.001371575, 0.001755547, 0.001443259,
0.001647977, 0.002886186, 0.00429965, 0.002226, 0.005068018,
0.002392742, 0.010747391, 0.001948935, 0.014982166, 0.004788058,
0.005418707, 0.031635842, 0.013330765, 0.004290994, 0.008529259,
0.009236926, 0.002794691, 0.004381456, 0.005029592, 0.02178649,
0.017924634, 0.014964051, 0.012455305, 0.017578538, 0.003522298,
0.003995365, 0.002304488, 0.0076246, 0.002853387, 0.004211933,
0.003113751, 0.028071121, 0.014845754, 0.013223408, 0.004993331,
0.011854977, 0.00307663, 0.008828247, 0.005489159, 0.005821341,
0.008176441, 0.005847643, 0.008632582, 0.005255749, 0.009172938,
0.005514234, 0.008449149, 0.005600061, 0.014342186, 0.005054358,
0.010036212, 0.005694254, 0.00962091, 0.006520761, 0.010771743,
0.005989528, 0.027574999, 0.015720081, 0.015474263, 0.01363116,
0.016104659, 0.009909327, 0.018895618, 0.006892656, 0.011912717,
0.004188733, 0.003491596, 0.004924819, 0.00164198, 0.010121838,
0.014226956, 0.007660564, 0.002976035, 0.005713842, 0.019527251,
0.007317763, 0.039021655, 0.004191221, 0.008165457, 0.002657193,
0.002549085, 0, 0.00068218, 0.002252117, 0.00795954, 0.005629613,
0.005016125, 0.002790605, 0.0084233, 0.006397561, 0.02614528,
0.025029065, 0.018955912, 0.003711371, 0.014559335, 0.004751886,
0.015360884, 0.009555682, 0.009930083, 0.004168767, 0.006446649,
0.002351582, 0.007755967, 0.00180232, 0.010208893, 0.010828096,
0.019613881, 0.002060375, 0.009214464, 0.001909286, 0.005662194,
0.0018259, 0.005803247, 0.002112633, 0.00328676, 0.001935102,
0.004649152, 0.002166196, 0.004801268, 0.001954427, 0.033229958,
0.019154351, 0.005806449, 0.00259939, 0.004925562, 0.002736231,
0.01016808, 0.002615967, 0.007851256, 0.003662972, 0.01091895,
0.002805302, 0.007332856, 0.030372904, 0.017612059)), row.names = c(NA,
142L), class = "data.frame")
CodePudding user response:
There are some NA's in your updated example dataset. If you remove the NAs, this approach works as expected:
library(tidyverse)
sdNDVI %>%
na.omit() %>%
mutate(across(3:5, ~ifelse(.x == min_sd, 1, 0))) %>%
pivot_longer(c(3:5)) %>%
group_by(target, name) %>%
summarise(count = sum(value)) %>%
ggplot(aes(x = name, fill = target, y = count))
geom_col(width = 0.25, position = position_dodge(width = 0.4))
#> `summarise()` has grouped output by 'target'. You can override using the
#> `.groups` argument.
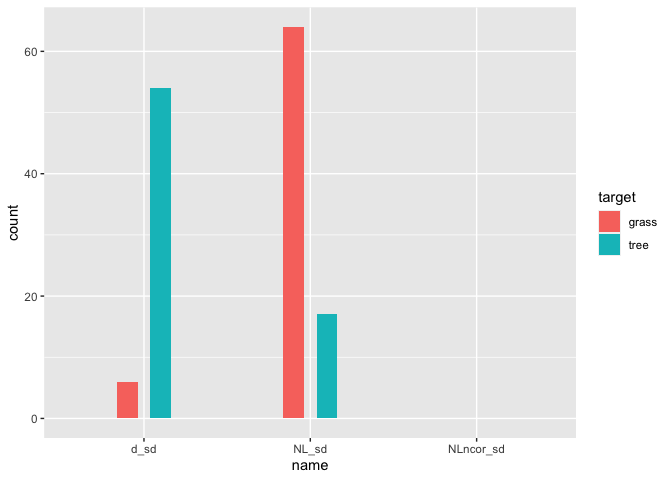
Created on 2022-07-26 by the reprex package (v2.0.1)