I'm new to R and coding in general and would really appreciate some help.
Say I have a dataframe, df1, with deveral columns. The important column is called Col1, the rest don't matter. This column contains only two classes: Yes or No
Something like this:
Col1
person1 Yes
person2 Yes
person3 No
person4 Yes
person5 No
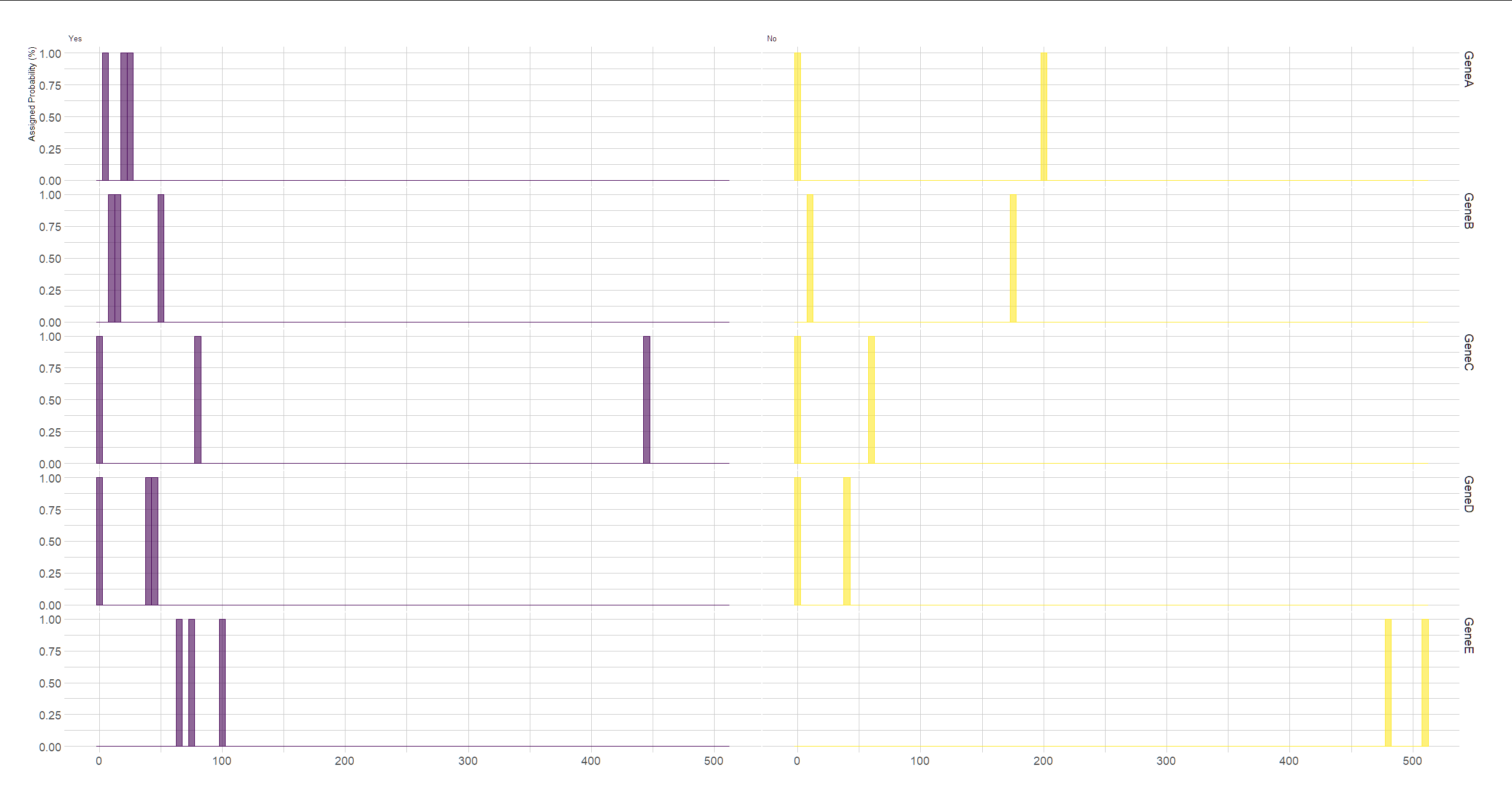
Another dataframe, df2, has those same samples in the columns, and genes in the rows. Something like this:
person1 person2 person3 person4 person5
GeneA 23 20 2 3 200
GeneB 9 50 11 17 177
GeneC 80 0 0 445 59
GeneD 43 39 38 0 1
GeneE 67 74 512 102 479
I want to make two histograms for each gene. One histogram would be only according to the samples with the label Yes in df1, and the second histogram would be for the same gene, but only for the samples that have the label No in df1. If it's possible, it would be even better if the two histograms were in the same plot, but with different colors, that would be clearer and easier to look at, to distinguish the difference between the two groups, in each gene.
How can I do that? Thank you very much.
CodePudding user response:
Something like this?
library(tidyverse)
library(viridis)
df1 <- df1 %>%
rownames_to_column()
df2 %>%
rownames_to_column() %>%
pivot_longer(-rowname) %>%
full_join(df1, by=c("name"="rowname")) %>%
mutate(Col1 = fct_reorder(Col1, value)) %>%
data.frame() %>%
ggplot(aes(x=value, color = Col1, fill = Col1))
geom_histogram(alpha=0.6, binwidth = 5)
scale_fill_viridis(discrete=TRUE)
scale_color_viridis(discrete=TRUE)
theme_ipsum()
theme(
legend.position="none",
panel.spacing = unit(0.1, "lines"),
strip.text.x = element_text(size = 8)
)
xlab("")
ylab("Assigned Probability (%)")
facet_grid(rowname~Col1)
data:
df1 <- structure(list(rowname = c("person1", "person2", "person3", "person4",
"person5"), Col1 = c("Yes", "Yes", "No", "Yes", "No")), row.names = c(NA,
-5L), class = "data.frame")
df2 <- structure(list(person1 = c(23L, 9L, 80L, 43L, 67L), person2 = c(20L,
50L, 0L, 39L, 74L), person3 = c(2L, 11L, 0L, 38L, 512L), person4 = c(3L,
17L, 445L, 0L, 102L), person5 = c(200L, 177L, 59L, 1L, 479L)), class = "data.frame", row.names = c("GeneA",
"GeneB", "GeneC", "GeneD", "GeneE"))
CodePudding user response:
We may use
library(dplyr)
library(tidyr)
library(ggplot2)
df2 %>%
rownames_to_column('rn') %>%
pivot_longer(cols = -rn) %>%
left_join(df1 %>%
rownames_to_column('name')) %>%
ggplot(aes(x = Col1, y = value, fill = Col1))
geom_col()
facet_wrap(~ rn, scales = "free")