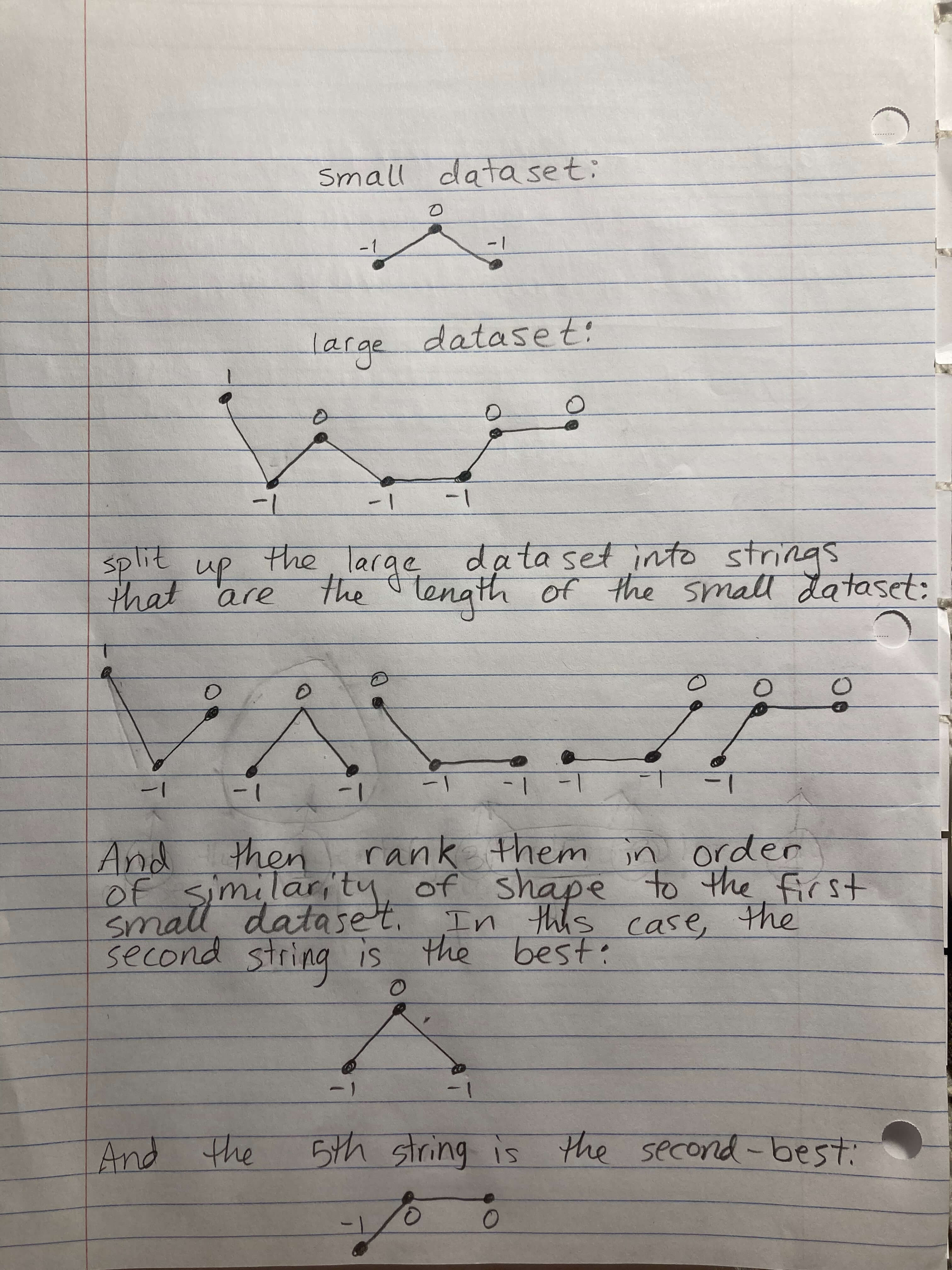
I am interested in taking a small sequence of numbers, for instance: -1, 0, -1.
And then looking within a larger dataset to find the most similar sequence of numbers within it. For example, the larger dataset could be: 1, -1, 0, -1, -1, 0, 0
The most similar sequence within it would be: 1, -1, 0, -1, -1, 0, 0
I believe the best strategy is to separate the larger dataset into several strings of the same length as the smaller dataset, in this case a length of 3, and then compare the smaller dataset to each of these strings and find the ones with the highest correlation. I would like to know which one is the closest, second-closest, third-closest, etc.
One key thing, I'm interested in which string has the most similar shape visually.
Please see my image below for a visualization of what I'm looking for:
I am a beginner, so if you could write out the code for me I would hugely appreciate it.
By the way, I am hoping to apply this function to much larger datasets than the one in this example.
Thank you!
CodePudding user response:
It's not clear what format you would like the answer in. Also, the notion of closest "shape" of data is too vague to encode. There are too many ways to interpret this. A simple Euclidean distance between the shorter vector and chunks of the longer vector of the same length makes most sense mathematically. You could code that like this:
closest_match <- function(needle, haystack) {
ln <- length(needle)
dist <- sapply(seq(length(haystack) - ln 1) - 1, function(i) {
sqrt(sum((haystack[i seq(ln)] - needle)^2))
})
list(index = which.min(dist),
closest_sequence = haystack[which.min(dist) seq(ln) -1])
}
And test it using your example vectors.
closest_match(c(-1, 0, -1), c(1, -1, 0, -1, -1, 0, 0))
#> $index
#> [1] 2
#>
#> $closest_sequence
#> [1] -1 0 -1
Here, index is the index of the long vector where the best match starts and closest_sequence is the actual best-fitting sequence within the longer vector.
Created on 2022-08-27 with reprex v2.0.2