My dataframe looks like this:
dput(cREP)
structure(list(date = structure(c(19165, 19166, 19166, 19167,
19167, 19168, 19168, 19169, 19169, 19170, 19170, 19171, 19171,
19172, 19172, 19173, 19173, 19174, 19174, 19175, 19175, 19176,
19176, 19177, 19177, 19178, 19178, 19179, 19179, 19180, 19180,
19181, 19181, 19182, 19182, 19183, 19183, 19184, 19184, 19185,
19185), class = "Date"), target = c("tree", NA, NA, "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", "grass", "tree", "grass", "tree", "grass",
"tree", "grass", "tree", NA, NA, NA, NA, "grass", "tree", "grass",
"tree", "grass", "tree"), sd_REP = c(0.151952193405258, NA, NA,
2.01826321634393, 0.169816578749757, 2.14229749687099, 0.28959682411733,
2.55083060426242, 0.295831565448322, 2.29505804254987, 0.576548440928348,
2.66666195619344, 0.292725343907804, 3.15812578193784, 2.18223221771057,
3.04511148118971, 0.252718667508799, 2.38208064405132, 0.292586777829021,
2.07630924668412, 0.338856357875883, 2.36807408753642, 0.282795351952376,
2.62641508244001, 0.225551429253523, 2.38246949685385, 0.295354071522136,
2.09719872313336, 0.291548177565861, 2.46644374339029, 0.31696780295781,
NA, NA, NA, NA, 2.35356881615488, 0.317570777241535, 2.72308512715765,
0.187766569086747, 1.9460851789847, 0.240891788076472), REP = c(724.650839520208,
NA, NA, 821.85192193247, 725.452721095604, 828.989588208101,
725.354782484439, 832.141025223582, 725.260289164661, 833.418421829249,
725.348412772341, 833.612432470951, 725.193602934028, 832.954790312524,
725.514259694303, 835.07931540604, 725.161575187682, 835.197801371358,
725.210328041105, 834.737425222639, 725.176653743869, 832.02623742477,
725.235318378951, 830.638384682061, 725.177856856095, 830.213290980754,
725.564603840649, 830.51494649741, 725.649417168378, 832.144511184231,
725.746812946587, NA, NA, NA, NA, 833.611415607686, 725.353584303352,
829.926258007964, 725.717456346066, 829.945696761203, 725.733792398208
), minTempAmb_Avg = c(NA, NA, NA, 13.93, NA, 14.31, NA, 12.78,
NA, 13.44, NA, 13.76, NA, 15.46, NA, 14.03, NA, 14.48, NA, 14.71,
NA, 15.96, NA, 15.37, NA, 14.22, NA, 14.6, NA, 19.26, NA, NA,
NA, NA, NA, 19.03, NA, 15.15, NA, 16.87, NA)), row.names = 150:190, class = "data.frame")
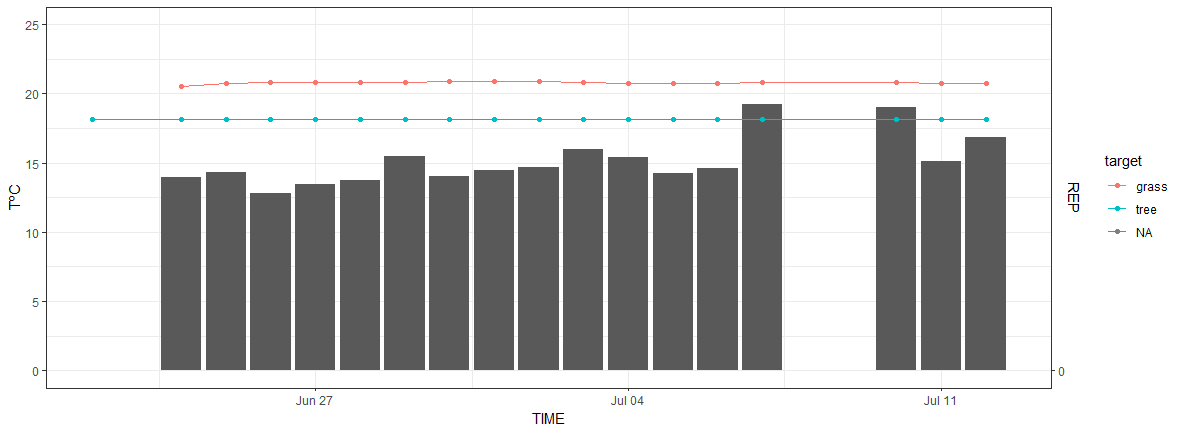
The two-way axis I'm plotting is the minimum temperature (minTempAmb_Avg geom_bar) with the REP (REP geom_line and geom_point) against time (date). I've successfully done the plot using this code:
cREP %>%
ggplot()
geom_bar(aes(x=date, y=minTempAmb_Avg),
stat="identity")
geom_point(aes(x=date, y=REP/40 , color=target),
stat="identity")
geom_line(aes(x=date, y=REP/40, color=target),
stat="identity")
geom_errorbar(aes(x = date,
y = REP ,
ymin=REP/40 -sd_REP/40,
ymax=REP/40 sd_REP/40,
width=0.01,
color = target
),
position = position_dodge(width = 0.05),
stat="identity")
labs(x="TIME", y="TºC")
scale_y_continuous(limits = c(0, 25), breaks=c(0, 5, 10, 15, 20, 25),
sec.axis = sec_axis(~.*1, name = "REP", breaks = seq(0, 400, 800)))
theme_bw()
This code is "shrinking" the REP data to the minTempAmb_Avg data and consequently does not allow us to see the standard deviations of REP. Plus I can't fit the REP scale on the right y-axis.
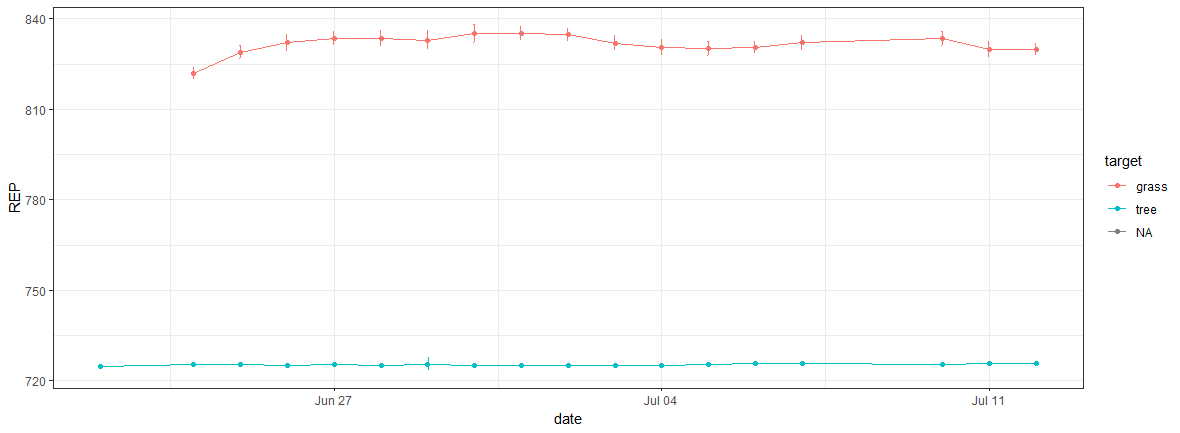
Check the plot of the REP without minTempAmb_Avg
The first thing to notice is the value discrepancy between minTempAmb_Avg and REP
range(cREP$REP, na.rm=T) #724.6508 835.1978
range(cREP$minTempAmb_Avg, na.rm=T) #12.78 19.26
How can I plot minTempAmb_Avg in the function of REP in a way I can see the standard deviations plus the scale in the right y-axis of REP?
Any help is much appreciated.
CodePudding user response:
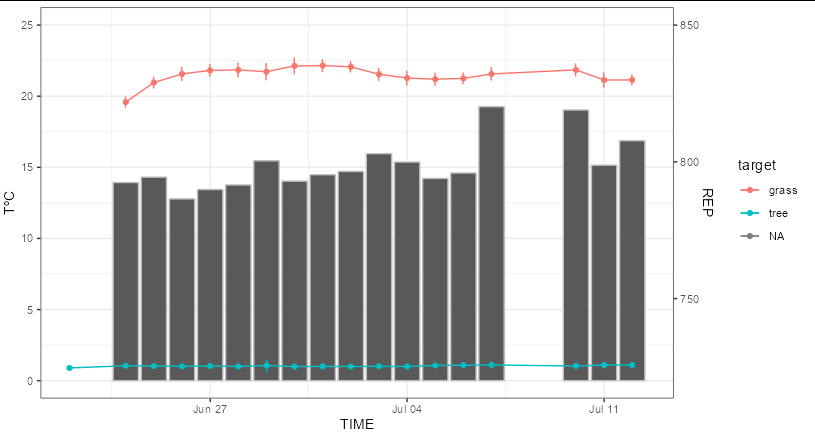
There are a couple of problems here. Firstly, you need to rescale REP so that both groups are maximally spread out on the y axis. Roughly speaking, this means you need to perform a transformation that maps the range [720, 850] onto the range [0, 25]. This would be achieved by the function (x - 720) / 5.2. Secondly, you need to transform sd_REP so that it is on the same scale, which would be sd_REP / 5.2. Lastly, you need the reverse transform on the secondary axis labels, which would be 5.2 * x 720
cREP %>%
mutate(REP = (REP - 720) / 5.2,
sd_REP = sd_REP / 5.2) %>%
ggplot(aes(x = date, y = REP, color = target))
geom_col(aes(y = minTempAmb_Avg), color = "gray")
geom_point(na.rm = TRUE)
geom_line(na.rm = TRUE)
geom_errorbar(aes(ymin = REP - sd_REP, ymax = REP sd_REP), width = 0.01,
position = position_dodge(width = 0.05), na.rm = TRUE)
labs(x = "TIME", y = "TºC")
scale_y_continuous(limits = c(0, 25), breaks=c(0, 5, 10, 15, 20, 25),
sec.axis = sec_axis(~720 .x * 5.2, name = "REP"))
theme_bw()