I have following cells:
cells = np.array([[1, 1, 1],
[1, 1, 0],
[1, 0, 0],
[1, 0, 1],
[1, 0, 0],
[1, 1, 1]])
and I want to calculate horizontal and vertical adjacencies to come to this result:
# horizontal adjacency
array([[3, 2, 1],
[2, 1, 0],
[1, 0, 0],
[1, 0, 1],
[1, 0, 0],
[3, 2, 1]])
# vertical adjacency
array([[6, 2, 1],
[5, 1, 0],
[4, 0, 0],
[3, 0, 1],
[2, 0, 0],
[1, 1, 1]])
The actual sollution looks like this:
def get_horizontal_adjacency(cells):
adjacency_horizontal = np.zeros(cells.shape, dtype=int)
for y in range(cells.shape[0]):
span = 0
for x in reversed(range(cells.shape[1])):
if cells[y, x] > 0:
span = 1
else:
span = 0
adjacency_horizontal[y, x] = span
return adjacency_horizontal
def get_vertical_adjacency(cells):
adjacency_vertical = np.zeros(cells.shape, dtype=int)
for x in range(cells.shape[1]):
span = 0
for y in reversed(range(cells.shape[0])):
if cells[y, x] > 0:
span = 1
else:
span = 0
adjacency_vertical[y, x] = span
return adjacency_vertical
The Algorithm is basically (for horizontal adjacency):
- loop throgh rows
- loop backward throgh columns
- if the x, y value of cells is not zero, add 1 to the actual span
- if the x, y value of cells is zero, reset actual span to zero
- set the span as new x, y value of the resulting array
Since I need to loop two times over all array elements this is slow for bigger arrays (e.g. images).
Is there a way to improve the algorithm using vectorization or some other numpy magic?
CodePudding user response:
I had a really quick attempt at this with Numba but have not checked it too thoroughly and have to go out:
#!/usr/bin/env python3
# https://stackoverflow.com/q/69854335/2836621
# magick -size 1920x1080 xc:black -fill white -draw "circle 960,540 960,1040" -fill black -draw "circle 960,540 960,800" a.png
import cv2
import numpy as np
import numba as nb
def get_horizontal_adjacency(cells):
adjacency_horizontal = np.zeros(cells.shape, dtype=int)
for y in range(cells.shape[0]):
span = 0
for x in reversed(range(cells.shape[1])):
if cells[y, x] > 0:
span = 1
else:
span = 0
adjacency_horizontal[y, x] = span
return adjacency_horizontal
@nb.jit('void(uint8[:,::1], int32[:,::1])',parallel=True)
def nb_get_horizontal_adjacency(cells, result):
for y in nb.prange(cells.shape[0]):
span = 0
for x in range(cells.shape[1]-1,0,-1):
if cells[y, x] > 0:
span = 1
else:
span = 0
result[y, x] = span
return
# Load image
im = cv2.imread('a.png', cv2.IMREAD_GRAYSCALE)
%timeit get_horizontal_adjacency(im)
result = np.zeros((im.shape[0],im.shape[1]),dtype=np.int32)
%timeit nb_get_horizontal_adjacency(im, result)
The timings are good, showing 4000x speed-up, if it works correctly:
In [15]: %timeit nb_get_horizontal_adjacency(im, result)
695 µs ± 9.12 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
In [17]: %timeit get_horizontal_adjacency(im)
2.78 s ± 44.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Input
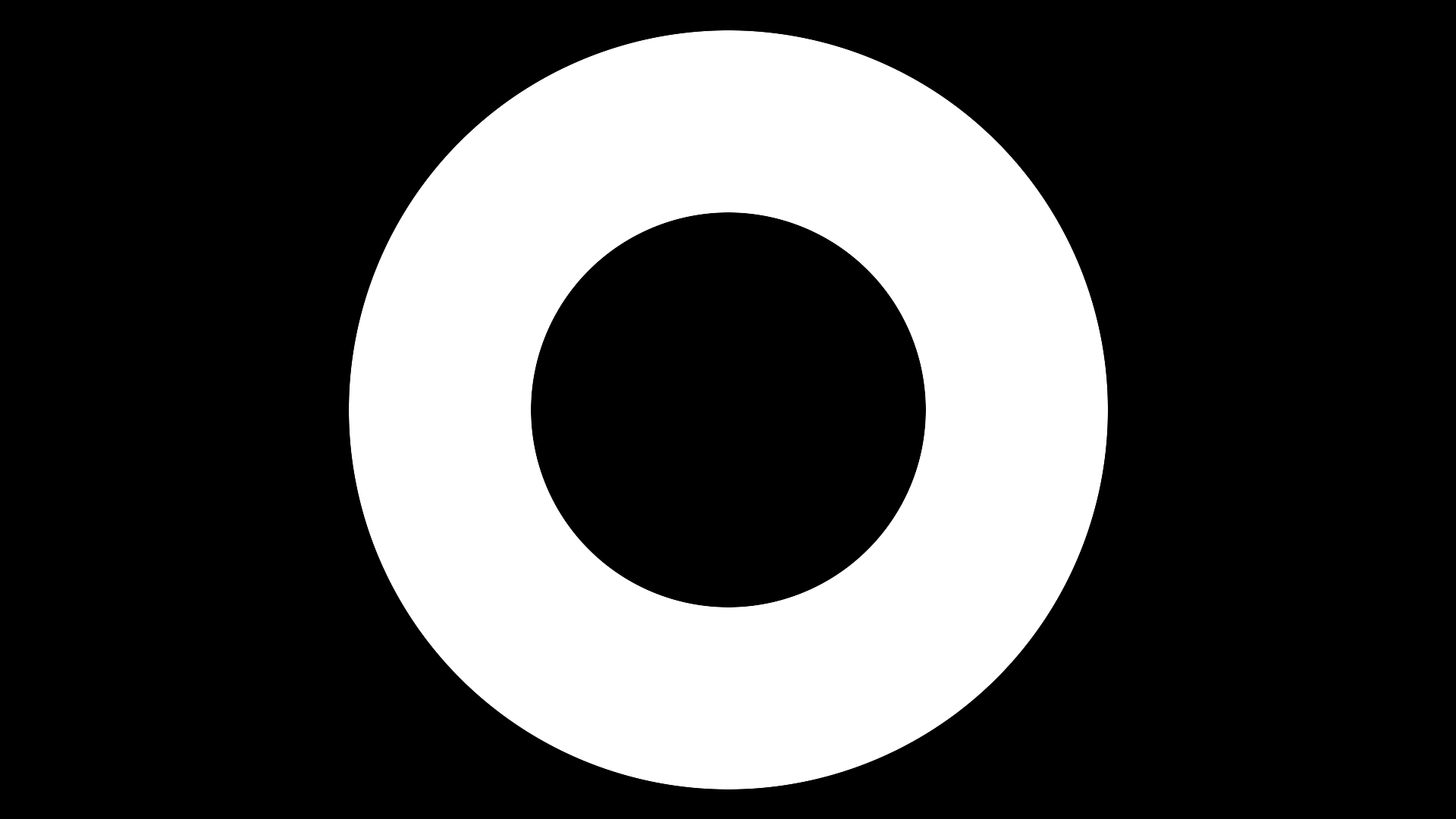
Input image was created in 1080p dimensions, i.e. 1920x1080, with ImageMagick using:
magick -size 1920x1080 xc:black -fill white -draw "circle 960,540 960,1040" -fill black -draw "circle 960,540 960,800" a.png
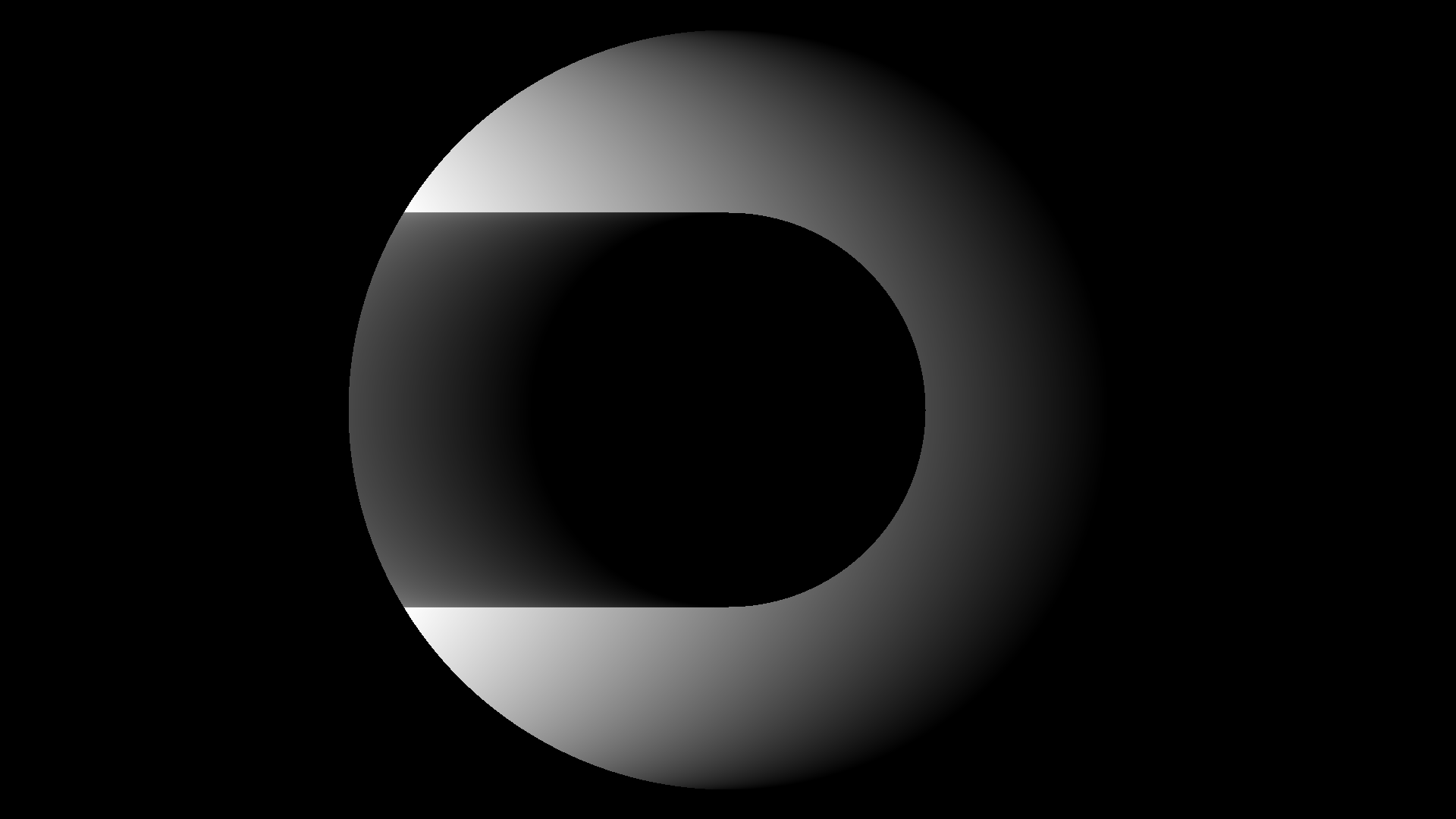
Output (contrast adjusted)
CodePudding user response:
As already stated in the comments, this is a perfect example where it's easier to just rewrite the function by means of Cython or Numba. Since Mark already provided a Numba solution, let me provide a Cython solution. First, let's time his solution on my machine for a fair comparison:
In [5]: %timeit nb_get_horizontal_adjacency(im, result)
836 µs ± 36 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
Assuming the image im is a np.ndarray with dtype=np.uint8, a parallelised Cython solution looks like this:
In [6]: %%cython -f -a -c=-O3 -c=-march=native -c=-fopenmp --link-args=-fopenmp
from cython import boundscheck, wraparound, initializedcheck
from libc.stdint cimport uint8_t, uint32_t
from cython.parallel cimport prange
import numpy as np
@boundscheck(False)
@wraparound(False)
@initializedcheck(False)
def cy_get_horizontal_adjacency(uint8_t[:, ::1] cells):
cdef int nrows = cells.shape[0]
cdef int ncols = cells.shape[1]
cdef uint32_t[:, ::1] adjacency_horizontal = np.zeros((nrows, ncols), dtype=np.uint32)
cdef int x, y, span
for y in prange(nrows, nogil=True, schedule="static"):
span = 0
for x in reversed(range(ncols)):
if cells[y, x] > 0:
span = 1
else:
span = 0
adjacency_horizontal[y, x] = span
return np.array(adjacency_horizontal, copy=False)
On my machine, this is nearly two times faster:
In [7]: %timeit cy_get_horizontal_adjacency(im)
431 µs ± 4.38 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)