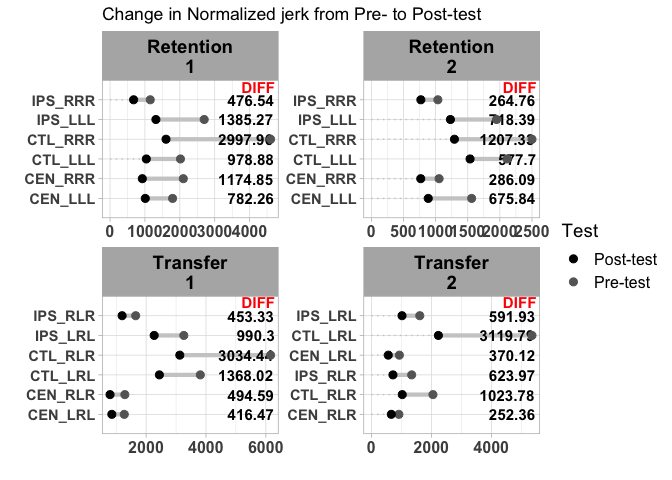
I tried plotting a dumbbell plot side by side with a facet grid but got some interesting errors. 
Error: At least one layer must contain all faceting variables: Trial_type, Session..
- Plot is missing
Trial_type,Session. - Layer 1 is missing
Session. - Layer 2 is missing
Session. - Layer 3 is missing
Session. - Layer 4 is missing
Session. - Layer 5 is missing
Trial_type,Session. - Layer 6 is missing
Session.Runrlang::last_error()to see where the error occurred. In addition: Warning messages: 1: In min(x) : no non-missing arguments to min; returning Inf 2: In max(x) : no non-missing arguments to max; returning -Inf 3: In min(x) : no non-missing arguments to min; returning Inf 4: In max(x) : no non-missing arguments to max; returning -Inf
data10A <- structure(list(Session = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2), conditon2 = structure(c(10L,
4L, 12L, 6L, 11L, 5L, 1L, 7L, 3L, 9L, 2L, 8L, 10L, 4L, 12L, 6L,
11L, 5L, 1L, 7L, 3L, 9L, 2L, 8L), .Label = c("CEN_LLL", "CTL_LLL",
"IPS_LLL", "CEN_RRR", "CTL_RRR", "IPS_RRR", "CEN_RLR", "CTL_RLR",
"IPS_RLR", "CEN_LRL", "CTL_LRL", "IPS_LRL"), class = "factor"),
Trial_type = structure(c(2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L,
1L, 2L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 2L, 1L, 2L, 1L,
2L), .Label = c("Retention", "Transfer"), class = "factor"),
NormalizedJerk_102 = c(1270.168699, 2099.703957, 3259.268053,
1152.257445, 3810.890123, 4601.918336, 1792.371775, 1288.768888,
2699.08162, 1650.968794, 2018.457394, 6159.567785, 931.350429,
1053.84252, 1611.673955, 1034.363607, 5352.195367, 2499.83996,
1560.678962, 915.3845866, 1948.757464, 1341.815274, 2113.732859,
2051.140838), NormalizedJerk_104 = c(853.7034116, 924.8554548,
2268.966702, 675.7160839, 2442.874632, 1603.954653, 1010.111276,
794.1752256, 1313.813984, 1197.638788, 1039.577947, 3125.131019,
561.2311988, 767.7541159, 1019.744071, 769.6067294, 2232.404471,
1292.509181, 884.8343164, 663.0273865, 1230.369444, 717.8466364,
1536.027898, 1027.358586), key = c("Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk", "Change in normalized jerk",
"Change in normalized jerk")), row.names = c(NA, -24L), class = c("tbl_df",
"tbl", "data.frame"))
library(tidyverse)
library(dumbbell)
dumbbell::dumbbell(data10A, id = "conditon2", key = "Trial_type",
leg = "Test",
column1 = "NormalizedJerk_102",
column2 = "NormalizedJerk_104",
delt = 1, lab1 = "Pre-test", lab2 = "Post-test",
p_col1 = "black", p_col2 = "grey40",
textsize = 4, segsize = 1.5,
pointsize = 2.5,
title = "Change in Normalized jerk from Pre- to Post-test")
facet_wrap( Trial_type ~ Session., scales="free", ncol=2)
theme(axis.text.x = element_text(size = 12, face = "bold"),
axis.text.y = element_text(size = 11, face = "bold"),
legend.position = "right",
legend.text = element_text(size = 12),
legend.title = element_text(size = 14),
strip.text = element_text(face = "bold", size = 14, color = "black"))
CodePudding user response:
The issue is that dumbbell::dumbbell does some data pre-processing before constructing the dumbbell chart via ggplot2. Among other things unnecessary columns get dropped during this step, i.e. only the columns specified in the arguments of dumbbell are kept while all other columns which in your case includes your Session column get dropped. As a consequence you are not able to facet by Session.
One workaround would be to add a new key column to your data which is simply the interaction of Trial_type and Session. This key column you could then passed to the key argument off dumbbell and could be used as faceting variable:
library(ggplot2)
library(dumbbell)
data10A$key <- paste(data10A$Trial_type, data10A$Session, sep = "\n")
dumbbell::dumbbell(data10A, id = "conditon2", key = "key",
leg = "Test",
column1 = "NormalizedJerk_102",
column2 = "NormalizedJerk_104",
delt = 1, lab1 = "Pre-test", lab2 = "Post-test",
p_col1 = "black", p_col2 = "grey40",
textsize = 4, segsize = 1.5,
pointsize = 2.5,
title = "Change in Normalized jerk from Pre- to Post-test")
facet_wrap( ~ key, scales="free", ncol=2)
theme(axis.text.x = element_text(size = 12, face = "bold"),
axis.text.y = element_text(size = 11, face = "bold"),
legend.position = "right",
legend.text = element_text(size = 12),
legend.title = element_text(size = 14),
strip.text = element_text(face = "bold", size = 14, color = "black"))
Created on 2021-11-14 by the reprex package (v2.0.1)
