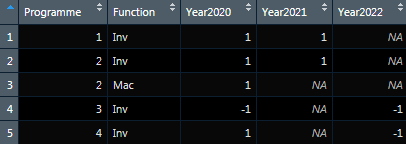
I have a list of columns where three of them relate to year, which identifies the occurrence of that data being collected for that year (1 or -1 means to be collected). I would like to create / manipulate the dataframe to have 'Year' as a new variable, which would replicate the other columns if it is to be collected for multiple years. What is the most efficient way to do this using dplyr / tidyverse?
I am aware of perhaps using pivot_longer but I assume I made need also to use case_when or similar. Any help appreciated.
Programme <- c(1, 2, 2, 3, 4)
Function <- c("Inv", "Inv", "Mac", "Inv", "Inv")
Year2020 <- c(1, 1, 1, -1, 1)
Year2021 <- c(1, 1, *NA*, *NA*, *NA*)
Year2022 <- c(*NA*, *NA*, *NA*, -1, -1)
df <- data.frame(Programme, Function, Year2020, Year2021, Year2022)
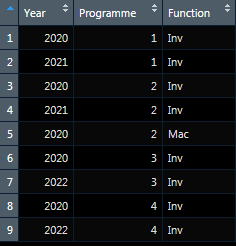
So what I am trying to produce is this:
Year <- c(2020, 2021, 2020, 2021, 2020, 2020, 2022, 2020, 2022)
Programme <- c(1, 1, 2, 2, 2, 3, 3, 4, 4)
Function <- c("Inv", "Inv", "Inv", "Inv", "Mac", "Inv", "Inv", "Inv", "Inv")
df <- data.frame(Year, Programme, Function)
CodePudding user response:
Using dplyr, tidyr and stringr packages:
library(tidyverse)
df |>
pivot_longer(3:5,
names_to = "Year") |>
filter(value == 1 | value == -1) |>
mutate(Year = str_remove(Year,"^Year")) |>
select(Year, Programme, Function)
Output:
# A tibble: 9 x 3
Year Programme Function
<chr> <dbl> <chr>
1 2020 1 Inv
2 2021 1 Inv
3 2020 2 Inv
4 2021 2 Inv
5 2020 2 Mac
6 2020 3 Inv
7 2022 3 Inv
8 2020 4 Inv
9 2022 4 Inv
CodePudding user response:
Update see comments Darren Tsai:"Adding names_transform = list(Year = parse_number) into pivot_longer() can save you the mutate line. Besides, in tidyr there is a more flexible function than na.omit() to treat NA, i.e. drop_na()"
library(tidyr)
library(dplyr)
df %>%
pivot_longer(
cols = starts_with("Year"),
names_to = "Year",
names_transform = list(Year = parse_number)
) %>%
drop_na() %>%
dplyr::select(Year, Programme, Function)
Alternative pivot method using names_pattern and regex "(\\D )(\\d )":
library(tidyr)
library(dplyr)
df %>%
pivot_longer(
cols = starts_with("Year"),
names_to = c(".value", "Year1"),
names_pattern = "(\\D )(\\d )"
) %>%
na.omit() %>%
dplyr::select(Year = Year1, Programme, Function)
OR using parse_number from readr package:
library(tidyverse)
df %>%
pivot_longer(
cols = starts_with("Year"),
names_to = "Year"
) %>%
mutate(Year = parse_number(Year)) %>%
na.omit() %>%
dplyr::select(Year, Programme, Function)
Year Programme Function
<chr> <dbl> <chr>
1 2020 1 Inv
2 2021 1 Inv
3 2020 2 Inv
4 2021 2 Inv
5 2020 2 Mac
6 2020 3 Inv
7 2022 3 Inv
8 2020 4 Inv
9 2022 4 Inv