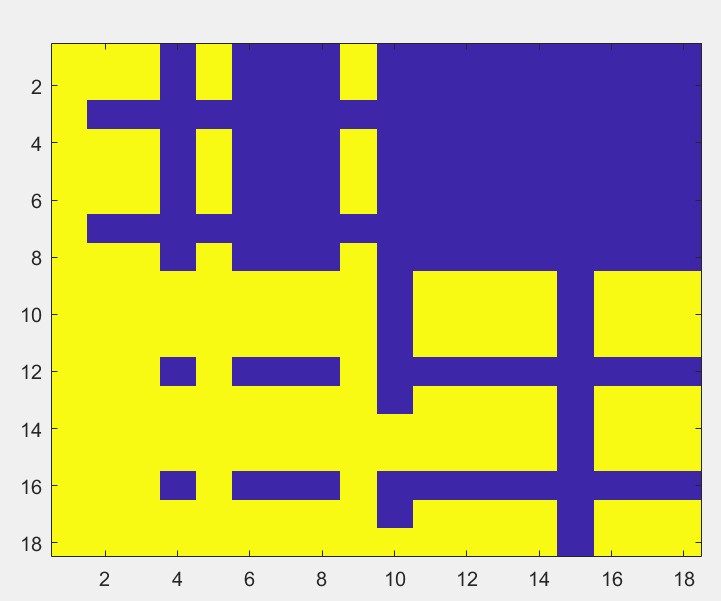
I have a xyz matrix stored in a .csv file that, for example, looks like the following:
1 2 3 4
1 1 1 -1 1
2 -1 1 1 1
3 1 -1 1 -1
4 1 1 -1 1
where the rows are x, columns are y and entries are z. I could store this matrix in a dataframe.
I want to create a heatmap of this matrix, where the entries would just be two colors, depending on 1 or -1. The plot would look like the following for an 18 X 18 matrix (done by a colleague in matlab probably):
where, say, yellow is for 1 and blue for -1.
How can I do this in R?
CodePudding user response:
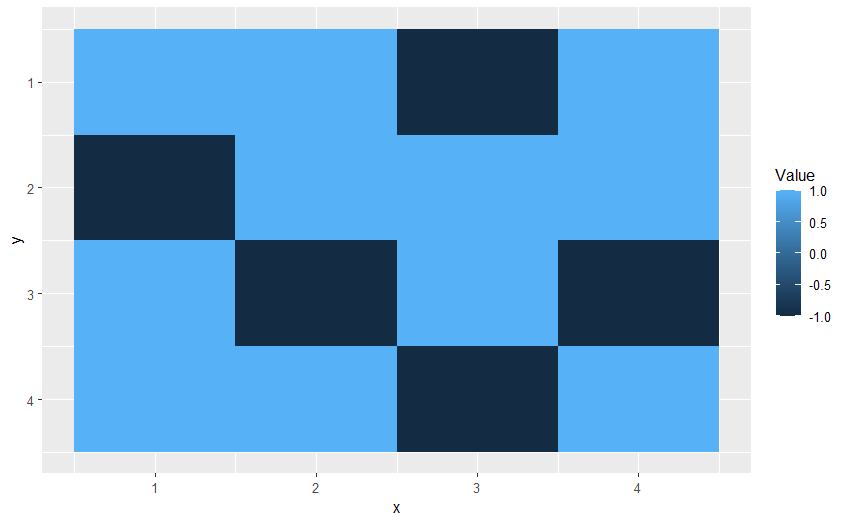
Use geom_tile(). First you need to flatten your dataframe using melt from the package reshape2.
library(reshape2)
flat_df <- melt(t(df), value.name = "Value")
ggplot(data=flat_df,aes(y=y,x=x)) geom_tile(aes(fill=Value)) scale_y_reverse()
CodePudding user response:
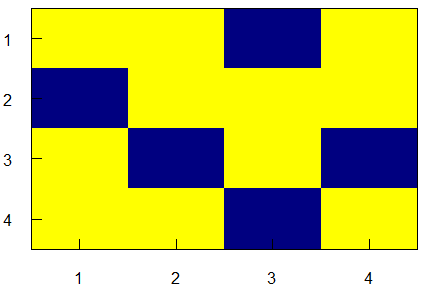
Multiple ways, but a base way is to use image(). Starting with a matrix:
# Matrix needs to be rotated 90 degrees before plotting with image()
m <- t(m[nrow(m):1, ])
image(m, axes = FALSE, col = c("navyblue", "yellow"))
axis(2, at = seq(0, 1, length.out = nrow(m)), labels = rev(seq(nrow(m))), las = 1, tcl = 0.5)
axis(1, at = seq(0, 1, length.out = ncol(m)), labels = seq(ncol(m)), tcl = 0.5)
box()
Data:
txt <- "1 1 -1 1
-1 1 1 1
1 -1 1 -1
1 1 -1 1"
m <- matrix(scan(text = txt, quiet = TRUE), byrow = TRUE, nrow = 4, dimnames = list(1:4, 1:4))