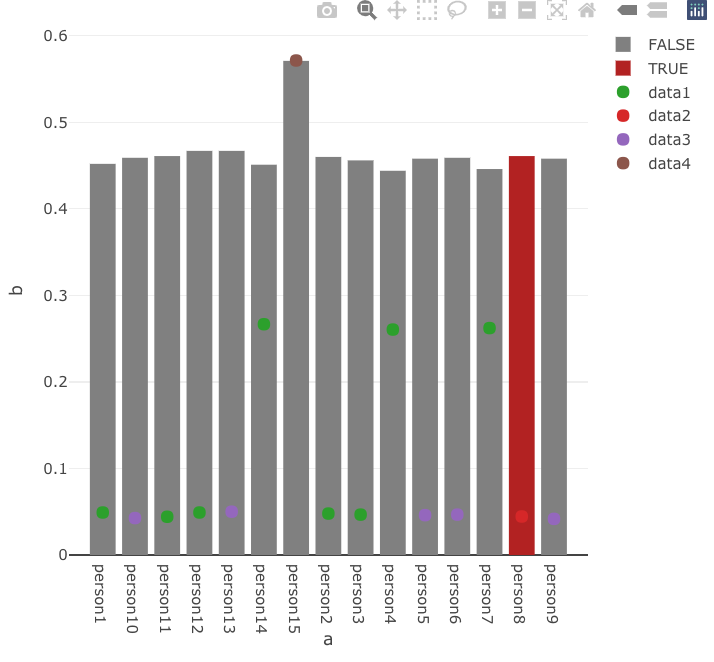
I'm currently developing a shiny application, and I started using Plotly for some graphs. The problem is that I want my barplot to be like that: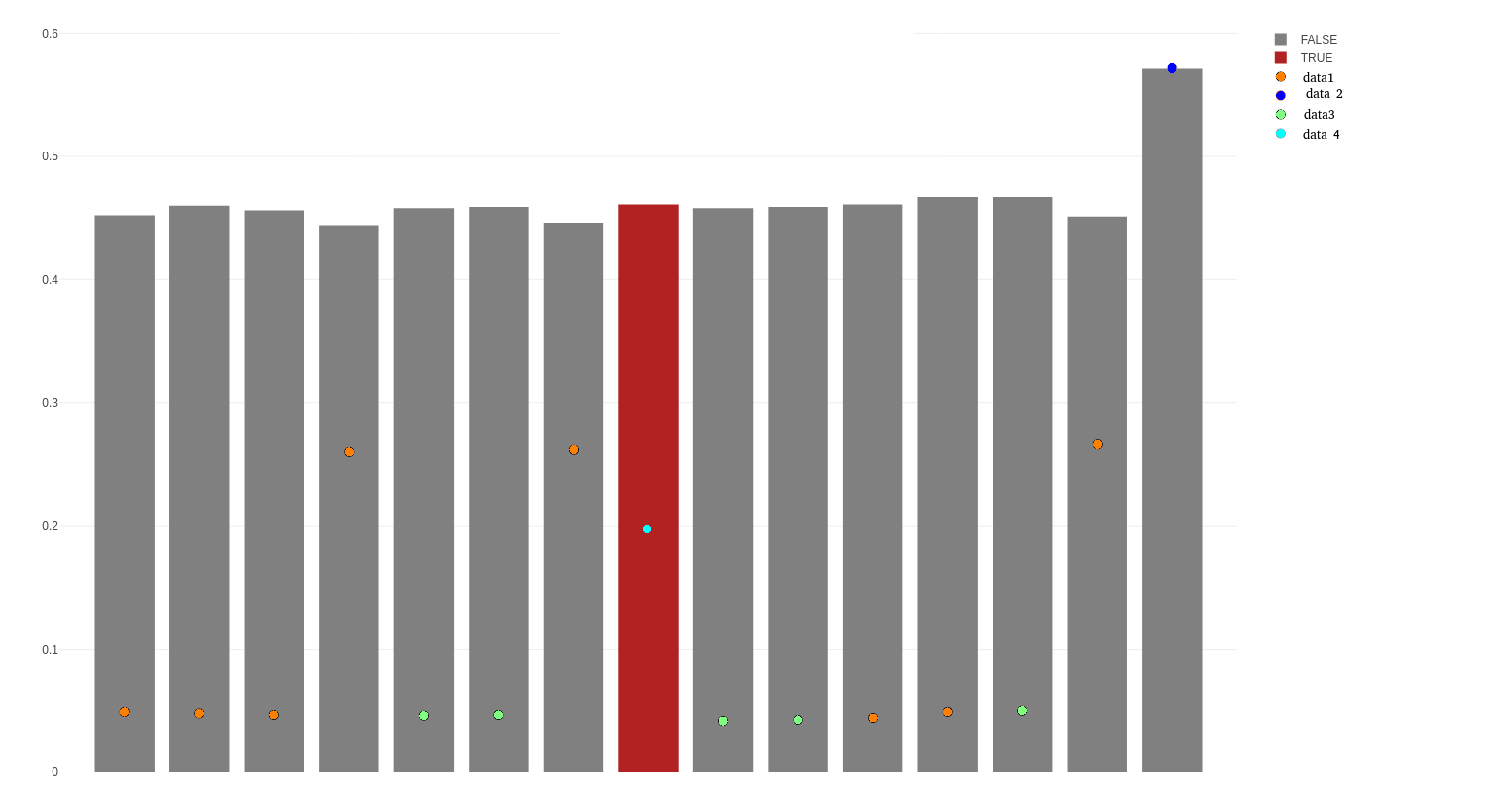
However, this is the resulting graph with the code I wrote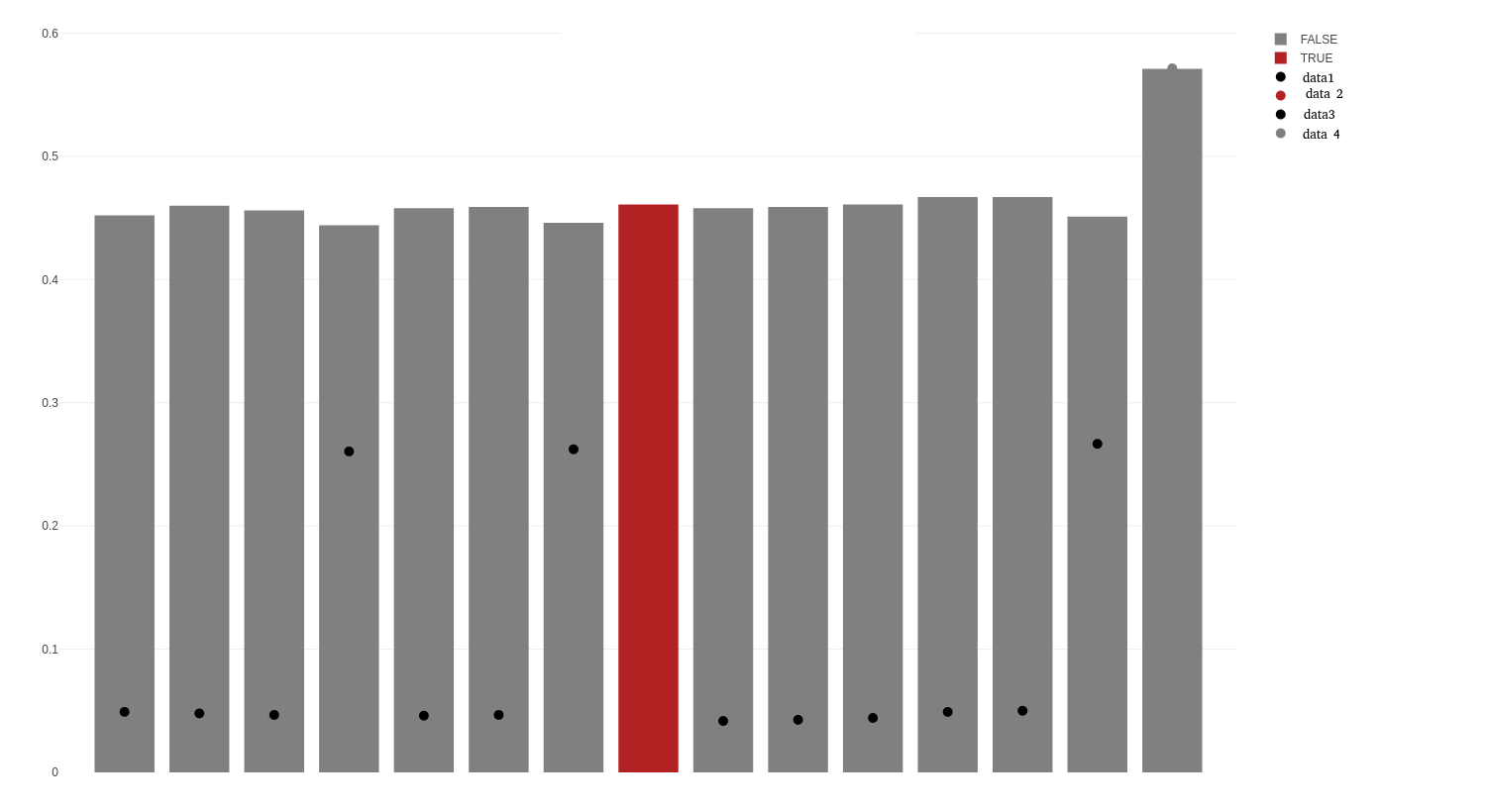
The code of the plot :
plot_ly(data =gfrq1, x= ~a, y= ~b, source = 'brand',type = 'bar',name = ~is.true, color = ~is.true, colors = c('TRUE' = "#B22222", 'FALSE' = '#808080'))%>%
add_trace(data = gfrq2, x = ~a, y= ~b, type = 'scatter', mode = 'marker point', name = ~data, marker = list( size =10 , color = ~data,colors = ~data))
The datasets are these :
dput(gfrq1)
structure(list(a = c("person1", "person2", "person3", "person4",
"person5", "person6", "person7", "person8", "person9", "person10",
"person11", "person12", "person13", "person14", "person15"),
b = c(0.452, 0.46, 0.456, 0.444, 0.458, 0.459, 0.446, 0.461,
0.458, 0.459, 0.461, 0.467, 0.467, 0.451, 0.571), is.true = c(FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, TRUE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE)), row.names = c(NA, -15L
), class = "data.frame")
dput(gfrq2)
structure(list(a = c("person1", "person2", "person3", "person4",
"person5", "person6", "person7", "person8", "person9", "person10",
"person11", "person12", "person13", "person14", "person15"),
b = c(0.049109883364027, 0.0478821362799263, 0.0466543891958257,
0.26051282051282, 0.0460405156537753, 0.0466543891958257,
0.262222222222222, 0.0445058317986495, 0.041743400859423,
0.0426642111724985, 0.0441988950276243, 0.049109883364027,
0.0500306936771025, 0.266666666666667, 0.571428571428571),
data = c("data1", "data1", "data1", "data1", "data3", "data3",
"data1", "data2", "data3", "data3", "data1", "data1", "data3",
"data1", "data4"), is.true = c(FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE)), row.names = c(NA, -15L), class = "data.frame")
Any suggestions?
CodePudding user response:
I was surprised how uncooperative Plotly was in this endeavor. Unfortunately, I have to admit that I stumbled upon the solution.
I joined the data into one data frame. Instead of using the parameter color for the markers, split instead.
I removed the parameter name from the add_bars, because with that set, the legend reflected "is.true" and "TRUE". (Why oh why can't Plotly read my mind?)
ggdat <- cbind(gfrq1, gfrq2[, c("b","data")]) # join data
names(ggdat) <- c("a", "b", "is.true", "b2", "dat") # fix names
plot_ly(data = ggdat, x = ~a) %>%
add_bars(y = ~b, color = ~is.true,
colors = c('TRUE' = "#B22222", 'FALSE' = '#808080')) %>%
add_markers(y = ~b2, split = ~dat, marker = list(size = 10))
I'm pretty sure there's no markers point mode. For dots, points, scatter...etc., the mode is markers. (I could be wrong, though!)