I have data of drug concentrations (DV) that include a parent drug and its metabolite, coded DVID 1 and 2, respectively. They are provided to me as separate rows, identified by study (STUDYID), subject (ID), and the time after the dose (TAD). I have created with paste an index column that I hope is unique to each study ID time. I would like to generate a data frame to plot the concentrations of the parent drug and metabolite against one another:
Index Drug Metabolite
Here are a few lines of data. Note that there is not a match for the drug and metabolite at time 0.5 and 1, so I would like to exclude these unpaired records. dcast wants an aggregation function - I just want to match up the two concentrations from the same sample. Also, can this be done without the concatenated Index column?
STUDYID ID TAD DV DVID Index
1 2 1 0.50000 0.80871 1 2SUBJ10.5
2 2 1 1.00000 2.53052 1 2SUBJ11
3 2 1 1.50000 2.64830 1 2SUBJ11.5
4 2 1 1.50000 -0.94495 2 2SUBJ11.5
5 2 1 2.01667 2.74019 1 2SUBJ12.01667
6 2 1 2.01667 -0.69375 2 2SUBJ12.01667
7 2 1 2.53333 2.91831 1 2SUBJ12.53333
8 2 1 2.53333 -0.30612 2 2SUBJ12.53333
9 2 1 3.00000 2.88926 1 2SUBJ13
10 2 1 3.00000 -0.12863 2 2SUBJ13
11 2 1 4.00000 2.78932 1 2SUBJ14
12 2 1 4.00000 0.01882 2 2SUBJ14
CodePudding user response:
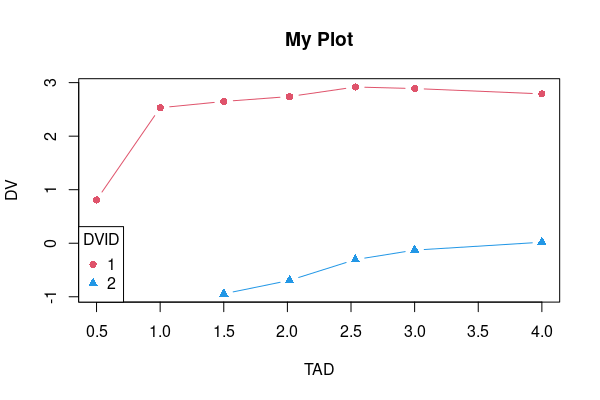
Not really sure what you want to dcast, you could simply plot the data directly.
plot(DV ~ TAD, df1, type='n', main='My Plot') ## initialize
lines(DV ~ TAD, df1[df1$DVID == 1, ], type='b', pch=16, col=2)
lines(DV ~ TAD, df1[df1$DVID == 2, ], type='b', pch=17, col=4)
legend('bottomleft', legend=1:2, title='DVID', col=c(2, 4), pch=16:17)
Data:
df1 <- structure(list(STUDYID = c(2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2
), ID = c(1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1), TAD = c(0.5, 1,
1.5, 1.5, 2.01667, 2.01667, 2.53333, 2.53333, 3, 3, 4, 4), DV = c(0.80871,
2.53052, 2.6483, -0.94495, 2.74019, -0.69375, 2.91831, -0.30612,
2.88926, -0.12863, 2.78932, 0.01882), DVID = c(1, 1, 1, 2, 1,
2, 1, 2, 1, 2, 1, 2), Index = c(" 2SUBJ10.5", " 2SUBJ11", " 2SUBJ11.5",
" 2SUBJ11.5", "2SUBJ12.01667", "2SUBJ12.01667", "2SUBJ12.53333",
"2SUBJ12.53333", " 2SUBJ13", " 2SUBJ13", " 2SUBJ14", " 2SUBJ14"
)), class = "data.frame", row.names = c(NA, -12L))
CodePudding user response:
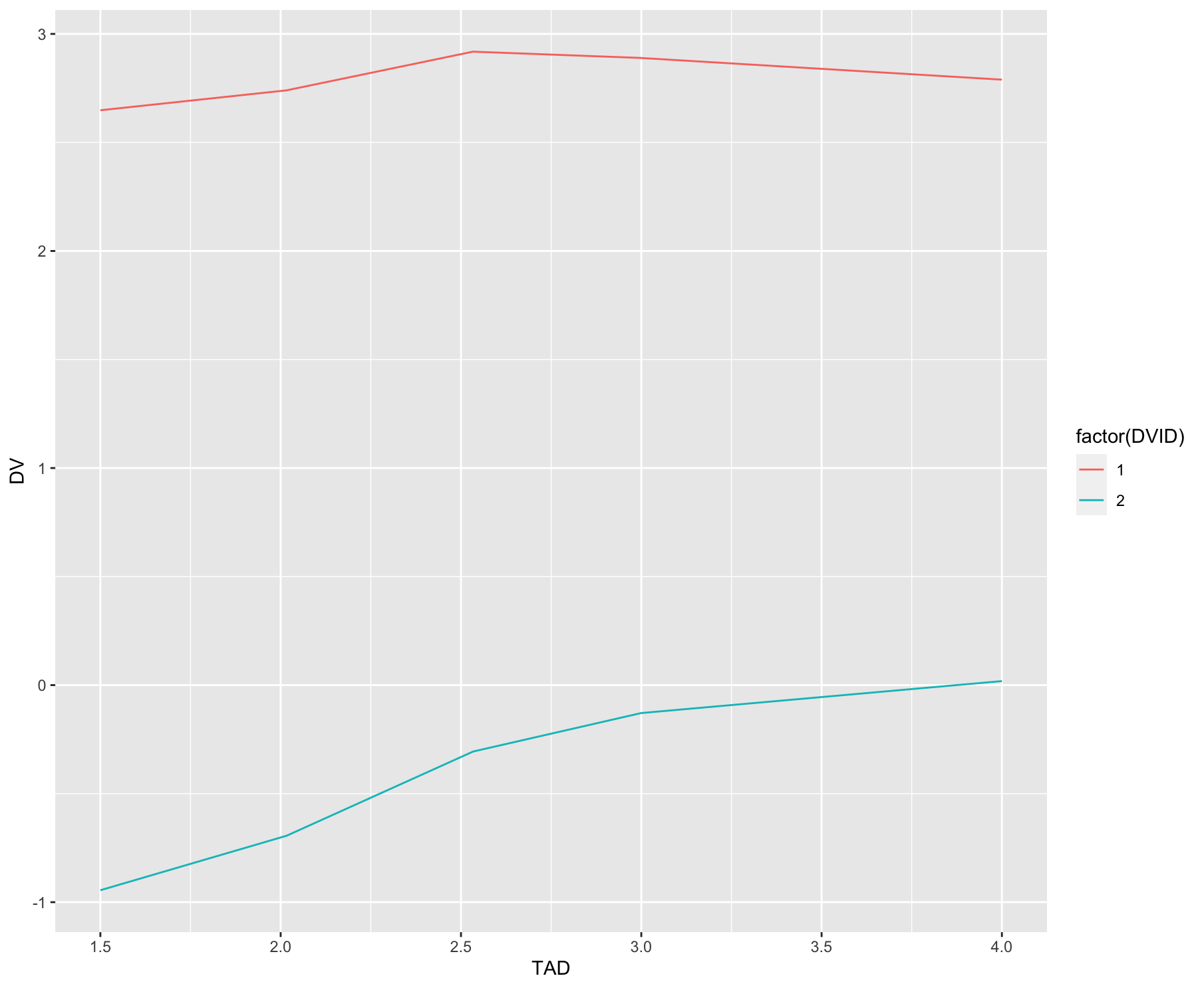
You can first remove the rows that are unpaired, then we can plot using ggplot.
library(tidyverse)
df %>%
group_by(STUDYID, ID, TAD) %>%
filter(n() > 1) %>%
ggplot()
geom_line(aes(x = TAD, y = DV, group = factor(DVID), color = factor(DVID)))
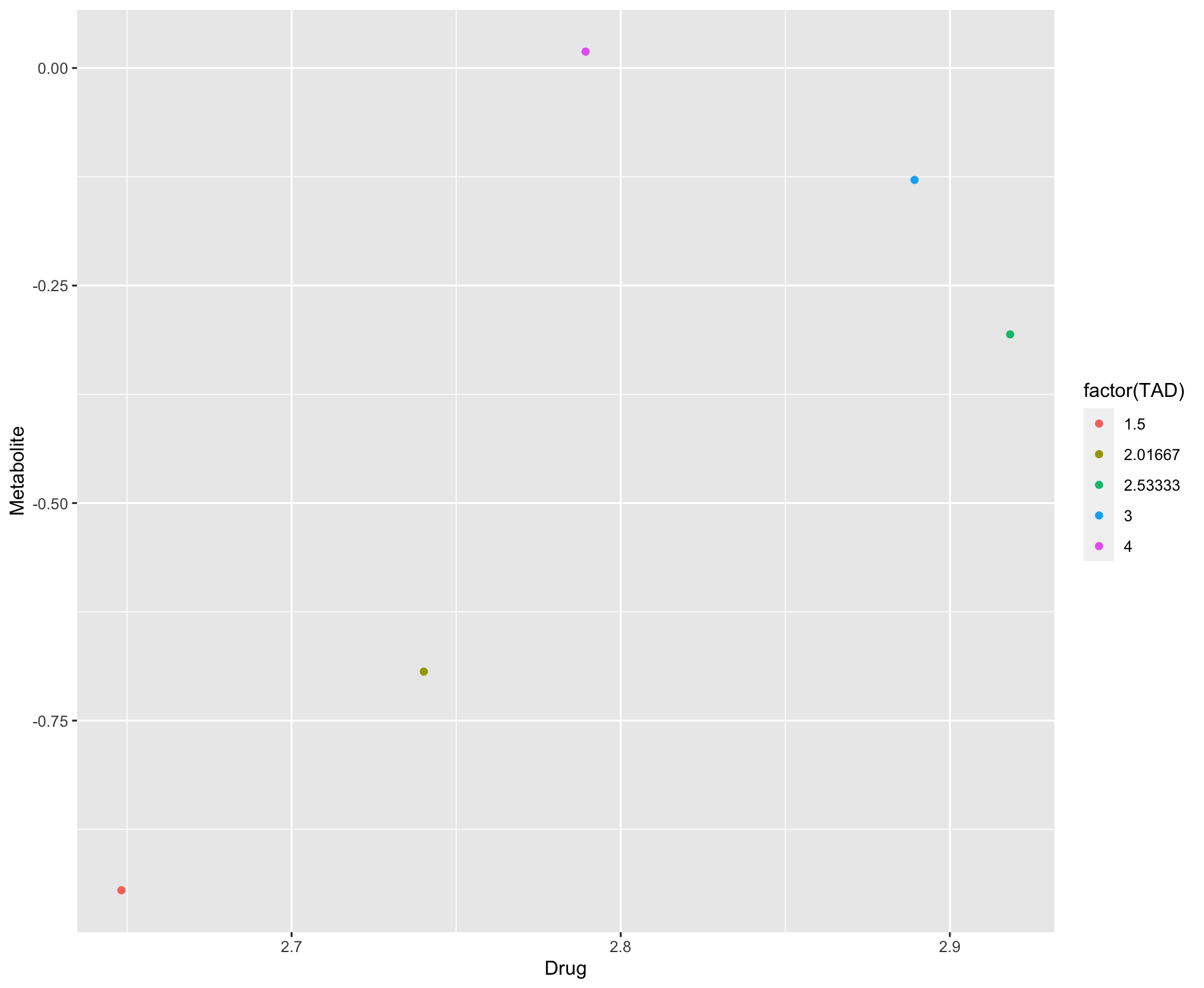
Alternatively, if you were wanting to plot the values of the pairs, then you could do something like this:
output <- df %>%
group_by(STUDYID, ID, TAD) %>%
filter(n() > 1) %>%
pivot_wider(names_from = "DVID", values_from = "DV") %>%
rename("Drug" = `1`, "Metabolite" = `2`)
ggplot(output)
geom_point(aes(x = Drug, y = Metabolite, color = factor(TAD)))
Data
df <- structure(list(STUDYID = c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L), ID = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L), TAD = c(0.5, 1, 1.5, 1.5, 2.01667, 2.01667, 2.53333, 2.53333,
3, 3, 4, 4), DV = c(0.80871, 2.53052, 2.6483, -0.94495, 2.74019,
-0.69375, 2.91831, -0.30612, 2.88926, -0.12863, 2.78932, 0.01882
), DVID = c(1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L),
Index = c("2SUBJ10.5", "2SUBJ11", "2SUBJ11.5", "2SUBJ11.5",
"2SUBJ12.01667", "2SUBJ12.01667", "2SUBJ12.53333", "2SUBJ12.53333",
"2SUBJ13", "2SUBJ13", "2SUBJ14", "2SUBJ14")), class = "data.frame", row.names = c(NA,
-12L))