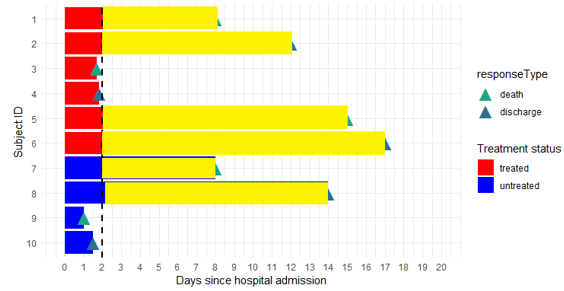
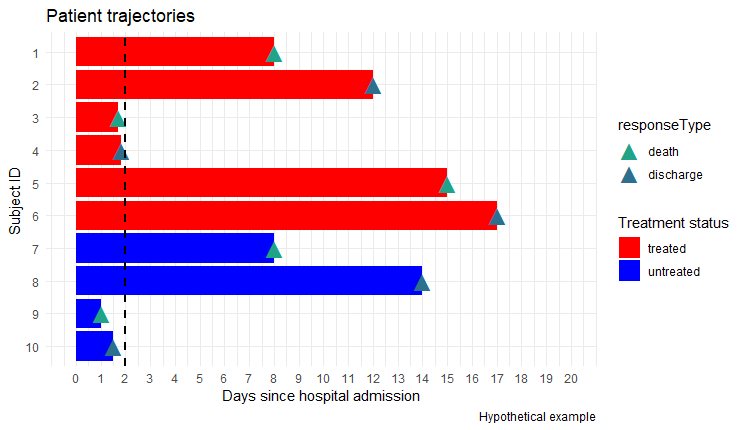
I'm wondering how can I change the color of the stacked bar after 2 days (geometric vertical line)?
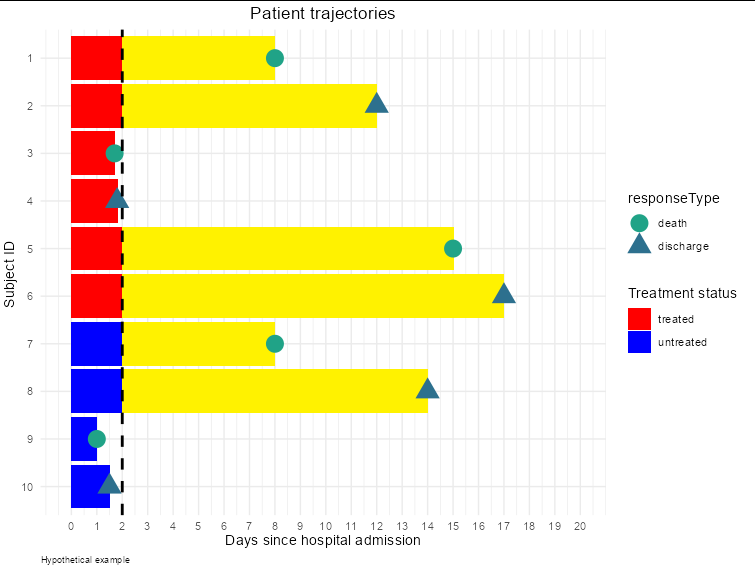
As a result, I want to get the figure marked with yellow color.
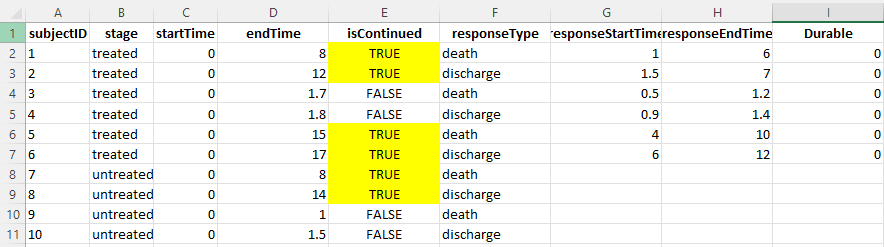
Here is an example from my data frame with the indicator (marked in yellow) if the subject continues after two days.
ggplot(aes(subjectID, endTime))
scale_fill_manual(values = c("red", "blue"))
geom_bar(stat="identity", aes(fill=factor(stage)))
geom_point(data=df, aes(subjectID, endTime, colour=responseType, shape=responseType), size=6)
coord_flip()
scale_colour_manual(values=c('#20A387FF','#2D708EFF'))
scale_y_continuous(limits=c(-0.2, 20), breaks=0:20)
labs(fill="Treatment status", # Add labels
x="Subject ID ", y="Days since hospital admission",
title="Patient trajectories",
caption="Hypothetical example")
geom_hline(yintercept=2, linetype="dashed", size=1) # , col=c('DC0000B2')
theme(plot.title = element_text(hjust = 0.5), # Put title in the middle of plot
plot.caption = element_text(size=7, hjust=0)) # Make caption size smaller
theme_minimal()
CodePudding user response:
I think I would do this with geom_rect
ggplot(df, aes(subjectID, endTime))
scale_fill_manual(values = c("red", "blue"))
geom_col(aes(fill = factor(stage)))
geom_rect(aes(ymin = 2, ymax = endTime, xmin = as.numeric(subjectID) - 0.45,
xmax = as.numeric(subjectID) 0.45), fill = '#fff200',
data = df[df$isContinued,])
geom_point(aes(colour = responseType, shape = responseType), size = 6)
coord_flip()
scale_colour_manual(values = c('#20A387FF','#2D708EFF'))
scale_y_continuous(limits = c(-0.2, 20), breaks = 0:20)
labs(fill = "Treatment status",
x = "Subject ID ",
y = "Days since hospital admission",
title = "Patient trajectories",
caption ="Hypothetical example")
geom_hline(yintercept = 2, linetype = "dashed", size = 1)
theme_minimal()
theme(plot.title = element_text(hjust = 0.5),
plot.caption = element_text(size = 7, hjust = 0))
Data in reproducible format
df <- data.frame(subjectID = factor(1:10, 10:1),
stage = rep(c("treated", "untreated"), times = c(6, 4)),
startTime = 0,
endTime = c(8, 12, 1.7, 1.8, 15,17, 8, 14, 1, 1.5),
isContinued = rep(c(!0, !1, !0, !1), times = c(2, 2, 4, 2)),
responseType = rep(c("death", "discharge"), 5))
df
#> subjectID stage startTime endTime isContinued responseType
#> 1 1 treated 0 8.0 TRUE death
#> 2 2 treated 0 12.0 TRUE discharge
#> 3 3 treated 0 1.7 FALSE death
#> 4 4 treated 0 1.8 FALSE discharge
#> 5 5 treated 0 15.0 TRUE death
#> 6 6 treated 0 17.0 TRUE discharge
#> 7 7 untreated 0 8.0 TRUE death
#> 8 8 untreated 0 14.0 TRUE discharge
#> 9 9 untreated 0 1.0 FALSE death
#> 10 10 untreated 0 1.5 FALSE discharge