I have a table with blood sample data collected over 3 period of times (Albumin1, Albumin2,Albumin3). Each sample belongs to a group (placebo vs. test, i.e. 1 or 2 in the table). The table looks like this ('File1'):
| Date1 | Albumin1 | Date2 | Albumin2 | Date3 | Albumine3 | Group |
|---|---|---|---|---|---|---|
| 20-11-21 | 21 | 06-12-21 | 21 | 08-03-22 | 28 | 1 |
| 26-11-21 | 19 | 16-12-21 | 23 | 07-03-22 | 26 | 2 |
| 26-11-21 | 31 | 16-12-21 | 24 | 08-03-22 | 29 | 2 |
| 27-11-21 | 31 | 16-12-21 | 23 | 09-03-22 | 26 | 2 |
| 26-11-21 | 32 | 16-12-21 | 25 | 08-03-22 | 28 | 1 |
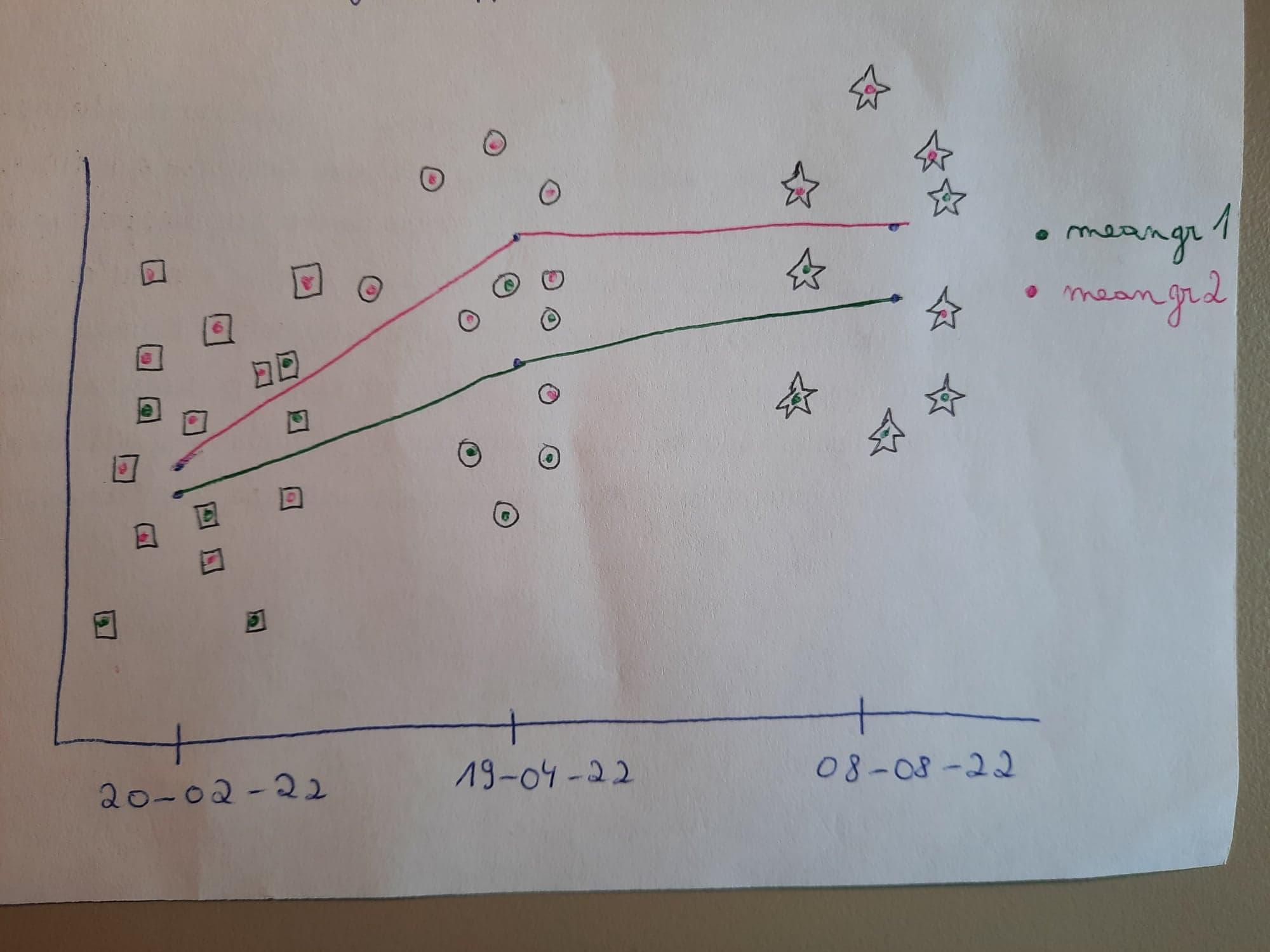
From this I would like to draw a time series plot like this, with colours per group and different symbols for the 3 series (as they can overlap) and with the average value for each group plotted with a line over time:
I assume that the first step involves the calculation of the means for each group and period of time. So I created a table ('Table2') summarising the data like this:
| Date_mean | Albumin_mean | Albumin_number | Group |
|---|---|---|---|
| 26-11-21 | 20 | 1 | 1 |
| 26-11-21 | 30 | 2 | 2 |
| 26-11-21 | 21 | 3 | 1 |
| 30-11-21 | 26 | 1 | 2 |
| 27-11-21 | 27 | 2 | 1 |
| 27-11-21 | 28 | 3 | 2 |
Here is the first attempt of ggplot code:
ggplot()
geom_point(data = Table1 , aes(x = as.Date(Date1,format=("%d-%m-%y")), aes(y=Albumin1), aes(colour = factor(Group)))
geom_line(data = Table2, aes(x = as.Date(Date_mean,format=("%d-%m-%y")), y=Albumin_mean))
I guess I have to add extra "geom_point()" for the 2 other scatter distributions, but I am stuck with the colours and symbols.
Many thanks
CodePudding user response:
It is difficult to get data in this format to plot nicely, so your first step should be to get your data into tidy format. This means each row should be an observation, and each column should be a variable. The date should also be in an actual Date format, rather than a string representation.
The following shows how we can get your table into this format:
library(tidyverse)
tidy_data <- Table1 %>%
pivot_longer(-Group, names_pattern = "(.*)(\\d)",
names_to = c('.value', "Series")) %>%
group_by(Series) %>%
mutate(Date = lubridate::dmy(Date),
Group = factor(Group))
We can see that we now have a single column for Albumin, a single column for Date, and columns to indicate which series and which group each measurement belongs to:
tidy_data
#> # A tibble: 15 x 4
#> # Groups: Series [3]
#> Group Series Date Albumin
#> <fct> <chr> <date> <int>
#> 1 1 1 2021-11-20 21
#> 2 1 2 2021-12-06 21
#> 3 1 3 2022-03-08 28
#> 4 2 1 2021-11-26 19
#> 5 2 2 2021-12-16 23
#> 6 2 3 2022-03-07 26
#> 7 2 1 2021-11-26 31
#> 8 2 2 2021-12-16 24
#> 9 2 3 2022-03-08 29
#> 10 2 1 2021-11-27 31
#> 11 2 2 2021-12-16 23
#> 12 2 3 2022-03-09 26
#> 13 1 1 2021-11-26 32
#> 14 1 2 2021-12-16 25
#> 15 1 3 2022-03-08 28
We can similarly make a tidy summary for each group / series combination like so:
summary_data <- tidy_data %>%
group_by(Series) %>%
mutate(Date = mean(Date)) %>%
group_by(Group, Date) %>%
summarize(Albumin = mean(Albumin))
summary_data
#> # A tibble: 6 x 3
#> # Groups: Group [2]
#> Group Date Albumin
#> <fct> <date> <dbl>
#> 1 1 2021-11-25 26.5
#> 2 1 2021-12-14 23
#> 3 1 2022-03-08 28
#> 4 2 2021-11-25 27
#> 5 2 2021-12-14 23.3
#> 6 2 2022-03-08 27
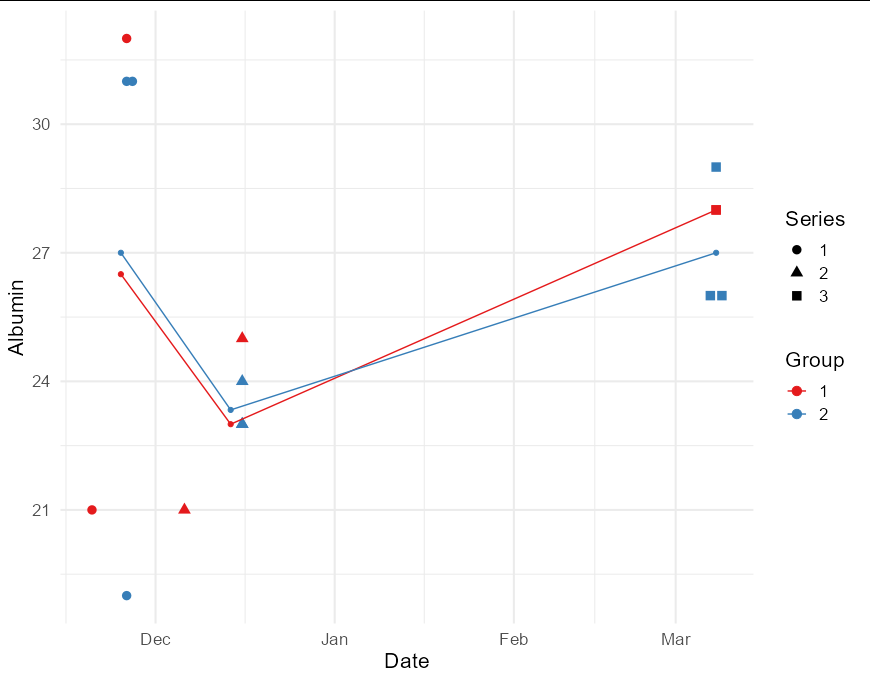
Now the plotting code itself is relatively straightforward:
ggplot(tidy_data, aes(Date, Albumin, color = Group))
geom_point(aes(shape = Series), size = 3)
geom_line(data = summary_data)
geom_point(data = summary_data)
scale_color_brewer(palette = "Set1")
theme_minimal(base_size = 16)
Created on 2022-08-05 by the reprex package (v2.0.1)
Data from question in reproducible format
Table1 <- structure(list(Date1 = c("20-11-21", "26-11-21", "26-11-21",
"27-11-21", "26-11-21"), Albumin1 = c(21L, 19L, 31L, 31L, 32L
), Date2 = c("06-12-21", "16-12-21", "16-12-21", "16-12-21",
"16-12-21"), Albumin2 = c(21L, 23L, 24L, 23L, 25L), Date3 = c("08-03-22",
"07-03-22", "08-03-22", "09-03-22", "08-03-22"), Albumin3 = c(28L,
26L, 29L, 26L, 28L), Group = c(1L, 2L, 2L, 2L, 1L)),
class = "data.frame", row.names = c(NA, -5L))