I'd like to add the significance letters to a plot using ggeffects package. In my case:
# Packages
library(ggeffects)
library(dplyr)
library(glmmTMB)
library(multcomp)
library(lsmeans)
library(ggplot2)
# My data set
ds <- read.csv("https://raw.githubusercontent.com/Leprechault/trash/main/temp_ger_ds.csv")
str(ds)
#'data.frame': 140 obs. of 4 variables:
# $ temp : chr "constante" "constante" "constante" "constante" ...
# $ generation : chr "G0" "G0" "G0" "G0" ...
# $ development: int 22 24 22 27 27 24 25 26 27 18 ...
General model:
mTCFd <- glmmTMB(development ~ temp * generation, data = ds,
family = ziGamma(link = "log"))
3 combinations plot:
#1) Plot for temp
lsm.mTCFd.temp <- lsmeans(mTCFd, c("temp"))
lt<-cld(lsm.mTCFd.temp, Letters=letters, decreasing = TRUE)
ds <- ds %>% mutate(x_1= 1 (readr::parse_number(generation)-2)*0.05,
group = generation)
df_gg <-ggpredict(mTCFd, terms = c("temp"))%>%
mutate(x_1= 1 (readr::parse_number(as.character(group))-2)*0.05)
df_gg %>% plot(add.data = TRUE)
geom_text(aes(x = x_1, label = lt[,7]), vjust = -0.5, show.legend = FALSE)
#2) Plot for generation
lsm.mTCFd.gera <- lsmeans(mTCFd, c("generation"))
lt<-cld(lsm.mTCFd.gera, Letters=letters, decreasing = TRUE)
ds <- ds %>% mutate(x_1= 1 (readr::parse_number(generation)-2)*0.05,
group = generation)
df_gg <-ggpredict(mTCFd, terms = c("generation"))%>%
mutate(x_1= 1 (readr::parse_number(as.character(group))-2)*0.05)
df_gg %>% plot(add.data = TRUE)
geom_text(aes(x = x_1, label = lt[,7]), vjust = -0.5, show.legend = FALSE)
#3) Plot for temp and generation interaction
lsm.mTCFd.temp.gera <- lsmeans(mTCFd, c("temp","generation"))
lt<-cld(lsm.mTCFd.temp.gera , Letters=letters, decreasing = TRUE)
ds <- ds %>% mutate(x_1= 1 (readr::parse_number(generation)-2)*0.05,
group = generation)
df_gg <-ggpredict(mTCFd, terms = c("temp","generation"))%>%
mutate(x_1= 1 (readr::parse_number(as.character(group))-2)*0.05)
df_gg %>% plot(add.data = TRUE)
geom_text(aes(x = x_1, label = lt[,8]), vjust = -0.5, show.legend = FALSE)
But I always as output:
Raw data not available.
Error in if (attr(x, "logistic", exact = TRUE) == "1" && attr(x, "is.trial", :
missing value where TRUE/FALSE needed
Please, any help with it?
CodePudding user response:
I think the code that is not working is mutate(), which drops all attributes from the data frame. This one works for me, when you replace the code-line that contains mutate():
lsm.mTCFd.temp <- lsmeans(mTCFd, c("temp"))
lt<-cld(lsm.mTCFd.temp, Letters=letters, decreasing = TRUE)
ds <- ds %>% mutate(x_1= 1 (readr::parse_number(generation)-2)*0.05,
group = generation)
# new lines, replace "mutate()" here
df_gg <-ggpredict(mTCFd, terms = c("temp"))
df_gg$x_1 <- 1 (readr::parse_number(as.character(df_gg$group))-2)*0.05
df_gg %>% plot(add.data = TRUE)
geom_text(aes(x = x_1, label = lt[,7]), vjust = -0.5, show.legend = FALSE)
CodePudding user response:
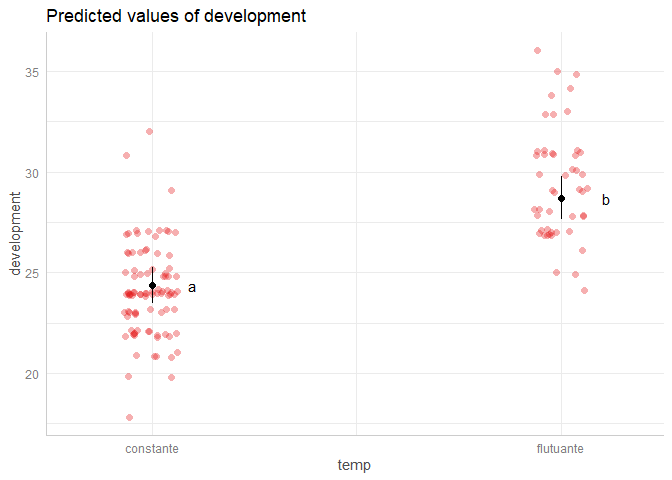
below is a reprex that takes your and @Daniel's code as its basis and simply moves the letters to the position you want. I only replaced {lsmeans} with its successor {emmeans} and edited the geom_text() argument a bit - thus, I did not have a closer look at what is actually done.
However, I suggest you read my answers to similar questions in the past here and with even more details here on specifically dealing with 2-way interactions emmeans. Both of them will also point you to this summary on compact letter displays. I hope this helps.
# Packages
library(ggeffects)
library(dplyr)
library(glmmTMB)
library(multcomp)
library(emmeans)
library(ggplot2)
# My data set
ds <- read.csv("https://raw.githubusercontent.com/Leprechault/trash/main/temp_ger_ds.csv")
# General Model
mTCFd <- glmmTMB(development ~ temp * generation,
data = ds,
family = ziGamma(link = "log"))
lsm.mTCFd.temp <- emmeans(mTCFd, c("temp"))
#> NOTE: Results may be misleading due to involvement in interactions
lt <- cld(lsm.mTCFd.temp, Letters = letters, decreasing = TRUE)
ds <- ds %>%
mutate(x_1 = 1 (readr::parse_number(generation) - 2) * 0.05, group = generation)
# new lines, replace "mutate()" here
df_gg <- ggpredict(mTCFd, terms = c("temp"))
df_gg$x_1 <-
1 (readr::parse_number(as.character(df_gg$group)) - 2) * 0.05
df_gg %>%
plot(add.data = TRUE)
geom_text(aes(x = x, label = lt[, 7]),
position = position_nudge(x = 0.1),
show.legend = FALSE)
Created on 2022-08-08 by the reprex package (v2.0.1)
