I'm trying to create a 3D shape plot, as is done in MATLAB using the patch command, like this one:
I've created my coordinate data using expand.grid:
library(pracma)
n1=60
n2=60
n3=60
data = expand.grid(x=linspace(-3,3,n1),y=linspace(-3,3,n2),z=linspace(-3,3,n3))
I've used this data to fill a 3D matrix to obtain a 3D matrix V, which defines the shape:
someData <- rep(0, 60*60*60)
V <- array(someData, c(60, 60, 60))
count = 1
lambda = c(2,1,1)
for (ii in 1:n1){
for (jj in 1:n2){
for (kk in 1:n3){
V[ii, jj, kk] = t(as.matrix(lambda))%*%as.matrix(c(data$x[count],data$y[count],data$z[count]))
count = count 1
}
}
}
However, I'm struggling to plot this in R.
Reproducible MATLAB example:
n1 = 60;
n2 = 60;
n3 = 60;
[y,x,z] = ndgrid(linspace(-3,3,n1),linspace(-3,3,n2),linspace(-3,3,n3));
V = zeros(n1, n2, n3);
lambda = [2;1;1];
for ii = 1: n1
for jj = 1:n2
for kk =1:n3
V(ii, jj, kk) = lambda'*[abs(y(ii, jj, kk)); abs(x(ii, jj, kk)); abs(z(ii, jj, kk))];
end
end
end
p = patch(isosurface(x,y,z,V,1));
p.FaceColor = 'cyan';
p.EdgeColor = 'none';
view(3);
camlight
axis equal
lighting gouraud
box on
My attempt in R:
n1=60
n2=60
n3=60
someData <- rep(0, 60*60*60)
V <- array(someData, c(60, 60, 60))
data = expand.grid(x=linspace(-3,3,n1),y=linspace(-3,3,n2),z=linspace(-3,3,n3))
lambda = c(2,1,1)
count = 1
for (ii in 1:n1){
for (jj in 1:n2){
for (kk in 1:n3){
V[ii, jj, kk] = t(as.matrix(lambda))%*%as.matrix(c(data$x[count],data$y[count],data$z[count]))
count = count 1
}
}
}
M = melt(V)
plot_ly(type = "mesh3d",
x = data$x,
y = data$y,
z = data$z,
i = M$Var1,
j = M$Var2,
k = M$Var3,
facecolor = rep(toRGB(colorRampPalette(c("navy", "blue"))(6)), each = 2)
)
However, I just get an empty coordinate axis, with no shape
CodePudding user response:
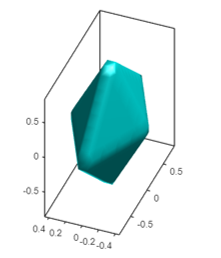
You can use the rmarchingcubes package to compute the isosurface and the rgl package to plot in 3D. The shape I get with the code below looks like a diamond. I didn't try to improve your code (there are possible improvements).
library(pracma)
library(rmarchingcubes) # to compute isosurface
library(rgl) # to plot in 3D
# your code with a couple of modifications
n1 <- 60
n2 <- 60
n3 <- 60
x <- linspace(-3, 3, n1)
y <- linspace(-3, 3, n2)
z <- linspace(-3, 3, n3)
data <- expand.grid(x = x, y = y, z = z)
someData <- rep(0, 60 * 60 * 60)
V <- array(someData, c(60, 60, 60))
count <- 1
lambda <- c(2, 1, 1)
for(ii in 1:n1) {
for(jj in 1:n2) {
for(kk in 1:n3) {
V[ii, jj, kk] <- t(as.matrix(lambda)) %*% # you forgot the absolute value:
abs(as.matrix(c(data$x[count], data$y[count], data$z[count])))
count <- count 1
}
}
}
# compute isosurface
isosurface <- contour3d(
griddata = V, level = 1, x = x, y = y, z = z
)
# make rgl mesh
mesh <- tmesh3d(
vertices = t(isosurface[["vertices"]]),
indices = t(isosurface[["triangles"]]),
normals = isosurface[["normals"]],
homogeneous = FALSE
)
# plot
shade3d(mesh, color = "turquoise")
EDIT
A big improvement to calculate V:
Mat <- abs(t(as.matrix(data)))
V <- array(t(lambda) %*% Mat, dim = c(n1, n2, n3))
EDIT
If you really want to use plotly:
library(plotly)
vs = isosurface[["vertices"]]
idx = isosurface[["triangles"]] - 1
plot_ly(
x = vs[,1], y = vs[,2], z = vs[,3],
i = idx[,1], j = idx[,2], k = idx[,3],
type = "mesh3d"
) %>% layout(scene = list(aspectmode = "data"))