I have multiple dataframes (sf objects). Each dataframe contains a geometry, name and probability column. They all have the same extents, but only overlap in some regions.
Here is some example data using two dataframes:
nc<-st_read(
system.file("gpkg/nc.gpkg",
package="sf"),
quiet=TRUE)
a<-nc %>%
select(c('geom')) %>%
slice(1:60) %>%
mutate(probability=runif(n=60,
min=1,
max=100)) %>%
mutate(name='A')
b<-nc %>%
select(c('geom')) %>%
slice(50:100) %>%
mutate(probability=runif(n=51,
min=1,
max=100)) %>%
mutate(name='B')
I would like to merge/join these two dataframes (a and b), but in the regions where they overlap, I would like to only keep the name where the probability is highest. The new dataframe should contain the name and probability.
How would I start with such a problem?
CodePudding user response:
In most cases you can handle sf objects as regular data.frames / tibbles with added benefits of spatial joins. Following example uses st_equals to limit matches to only those that originate from nc[50:60,] (neighbouring polygons in nc overlap, so the number of matches with st_intersects would be higher), for your real data you probably want to use something else (like default st_intersects), check ?st_join for alternatives.
library(dplyr)
library(sf)
library(ggplot2)
# by default st_join uses st_intersects predicate & left_join
# switching to st_equals & inner join
ab <- st_join(a, b, join = st_equals, suffix = c("_a", "_b"), left = F) %>%
mutate(
name = if_else(probability_a > probability_b, name_a, name_b),
probability = pmax(probability_a, probability_b)
# uncomment to only keep unused columns
# , .keep = "unused"
)
ggplot()
geom_sf(data = a, aes(alpha = probability), color = "gray80", fill = "red")
geom_sf(data = b, aes(alpha = probability), color = "gray80", fill = "green")
geom_sf(data = ab, aes(alpha = probability), color = "gray80", fill = "blue")
geom_sf_label(data = ab, aes(label = name), alpha = .5)
theme_void()
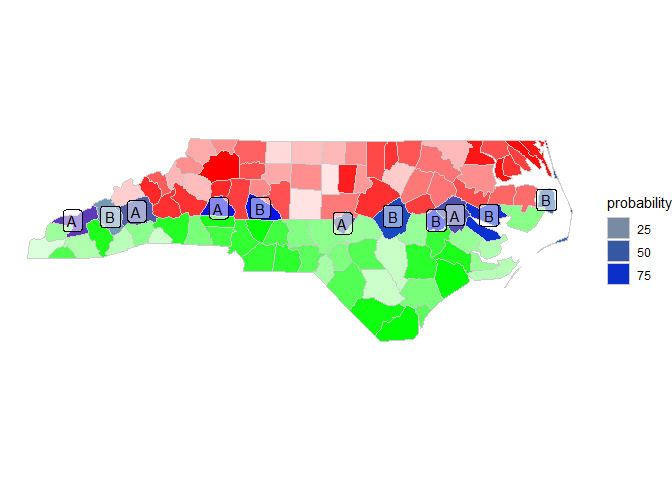 Resulting sf:
Resulting sf:
ab
#> Simple feature collection with 11 features and 6 fields
#> Geometry type: MULTIPOLYGON
#> Dimension: XY
#> Bounding box: xmin: -83.9547 ymin: 35.18983 xmax: -75.45698 ymax: 36.22926
#> Geodetic CRS: NAD27
#> First 10 features:
#> probability_a name_a probability_b name_b geom
#> 50 69.580424 A 91.374717 B MULTIPOLYGON (((-80.29824 3...
#> 51 48.284343 A 30.066734 B MULTIPOLYGON (((-77.47388 3...
#> 52 86.259738 A 46.447507 B MULTIPOLYGON (((-80.96143 3...
#> 53 44.371614 A 33.907073 B MULTIPOLYGON (((-82.2581 35...
#> 54 25.234930 A 65.436176 B MULTIPOLYGON (((-78.53874 3...
#> 55 7.997226 A 26.543661 B MULTIPOLYGON (((-82.74389 3...
#> 56 10.847150 A 48.375980 B MULTIPOLYGON (((-75.78317 3...
#> 57 32.310899 A 76.864756 B MULTIPOLYGON (((-77.10377 3...
#> 58 52.344792 A 9.340445 B MULTIPOLYGON (((-83.33182 3...
#> 59 66.538503 A 87.656812 B MULTIPOLYGON (((-77.80518 3...
#> name probability
#> 50 B 91.37472
#> 51 A 48.28434
#> 52 A 86.25974
#> 53 A 44.37161
#> 54 B 65.43618
#> 55 B 26.54366
#> 56 B 48.37598
#> 57 B 76.86476
#> 58 A 52.34479
#> 59 B 87.65681
Input:
set.seed(1)
nc <- st_read(
system.file("gpkg/nc.gpkg",
package = "sf"
),
quiet = TRUE
)
a <- nc %>%
select(c("geom")) %>%
slice(1:60) %>%
mutate(probability = runif(
n = 60,
min = 1,
max = 100
)) %>%
mutate(name = "A")
b <- nc %>%
select(c("geom")) %>%
slice(50:100) %>%
mutate(probability = runif(
n = 51,
min = 1,
max = 100
)) %>%
mutate(name = "B")
Created on 2022-11-21 with reprex v2.0.2
