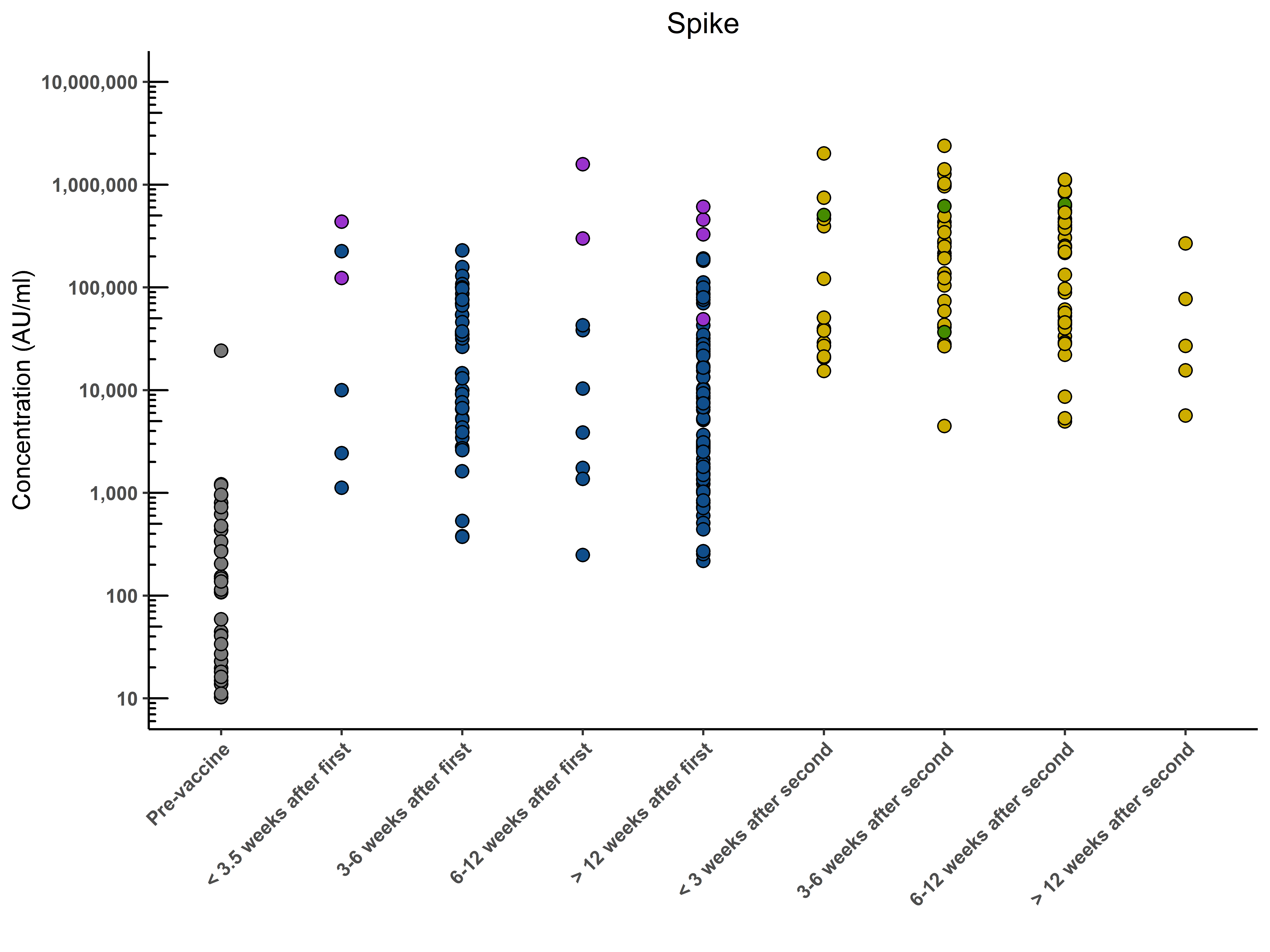
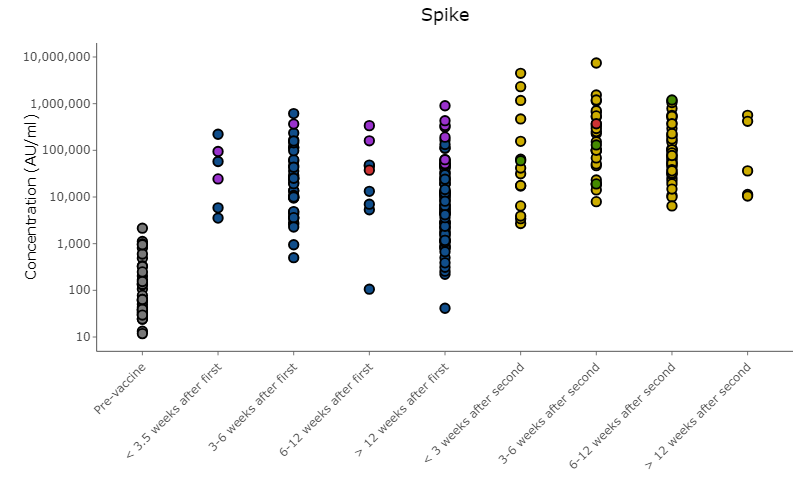
I want to send participants in my cohort their specific results shown graphically. There are over a hundred participants with multiple time points, the way I hope to send them is by colouring their specific data points red and keep the other sample IDs conditional on grouping (ie. vaccine status). I know how to colour points conditional on a group but am having trouble including another condition based on specific study IDs. The code below plugged into Plotly works seamlessly but Plotly also has its issues. I prefer to use ggplot2 to produce the graphs.
Unfortunately, I cannot share the dataset so here is a brief description:
- Sample = Study ID unique to each participant
- Timepoint = Blood sample collection relative to vaccination dates (first or second)
- Group = Condition describing the vaccine and infection status of each participant per timepoint (ie. Group 0 -> no vaccine ; 1 -> single dose; etc.)
Here's my code:
df %>%
mutate(Group = as.factor(Group)) %>%
mutate(Timepoint = fct_relevel(Timepoint, c("Pre-vaccine",
"< 3.5 weeks after first",
"3-6 weeks after first",
"6-12 weeks after first",
"> 12 weeks after first",
"< 3 weeks after second",
"3-6 weeks after second",
"6-12 weeks after second",
"> 12 weeks after second"))) %>%
droplevels() %>%
filter(Assay == "Antibody levels" & Group %in% c(0,1,2,3,4)) %>%
ggplot(aes(Timepoint, Concentration))
geom_jitter(position = position_jitter(width = 0.0001),
aes(fill = ifelse(str_detect(Sample, "V1"), Sample, Group)), # Here is where I specify the colour fill of data points if they match the study ID, if not they are coloured by 'Group'
pch = 21,
size = 2.5)
scale_y_log10(labels = scales::comma,
limits = c(10,10000000),
breaks = breaks,
minor_breaks = minor_breaks)
theme_classic()
labs(title = "Antibody levels",
x = "",
y = "Concentration (AU/ml)")
annotation_logticks(base = 10, sides = "l")
scale_fill_manual(values = pal)
theme(plot.title = element_text(hjust = 0.5),
axis.text.y = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1, face = "bold"),
legend.position = "none")
Figure 1. Example of colouring just by grouping. Ignores Sample ID.
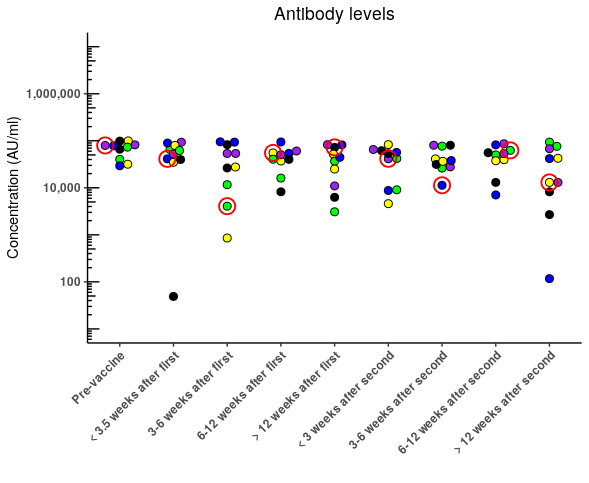
Figure 2. Figure produced in Plotly showing the desired outcome. Can this be done in ggplot2?
Edit: It seems to actually be working in ggplot but the red points are masked by the other data points. Is there a way to move them to the front while minimizing the amount of code?
CodePudding user response:
I will try to help you. But without your data I won't be able to do it exactly. So I generated data that was supposed to resemble your data. Notice one small detail of the Timepoint to factor variable with the labels attribute. This is a bit different than in your data.
library(tidyverse)
n=400
TimepointLev = c("Pre-vaccine",
"< 3.5 weeks after first",
"3-6 weeks after first",
"6-12 weeks after first",
"> 12 weeks after first",
"< 3 weeks after second",
"3-6 weeks after second",
"6-12 weeks after second",
"> 12 weeks after second")
df = tibble(
Sample = 1:n,
Group = sample(0:4, n, replace = TRUE) %>% paste() %>% factor(),
Timepoint = sample(1:9, n, replace = TRUE) %>% paste()
%>% factor(labels=TimepointLev),
Concentration = sample(1:15000, n, replace = TRUE)
)
df
output
# A tibble: 400 x 4
Sample Group Timepoint Concentration
<int> <fct> <fct> <int>
1 1 2 6-12 weeks after second 12021
2 2 1 > 12 weeks after second 13608
3 3 4 6-12 weeks after second 10417
4 4 0 3-6 weeks after second 2545
5 5 2 Pre-vaccine 2167
6 6 2 6-12 weeks after first 13725
7 7 3 3-6 weeks after second 3367
8 8 0 Pre-vaccine 3900
9 9 1 > 12 weeks after second 144
10 10 0 < 3 weeks after second 8219
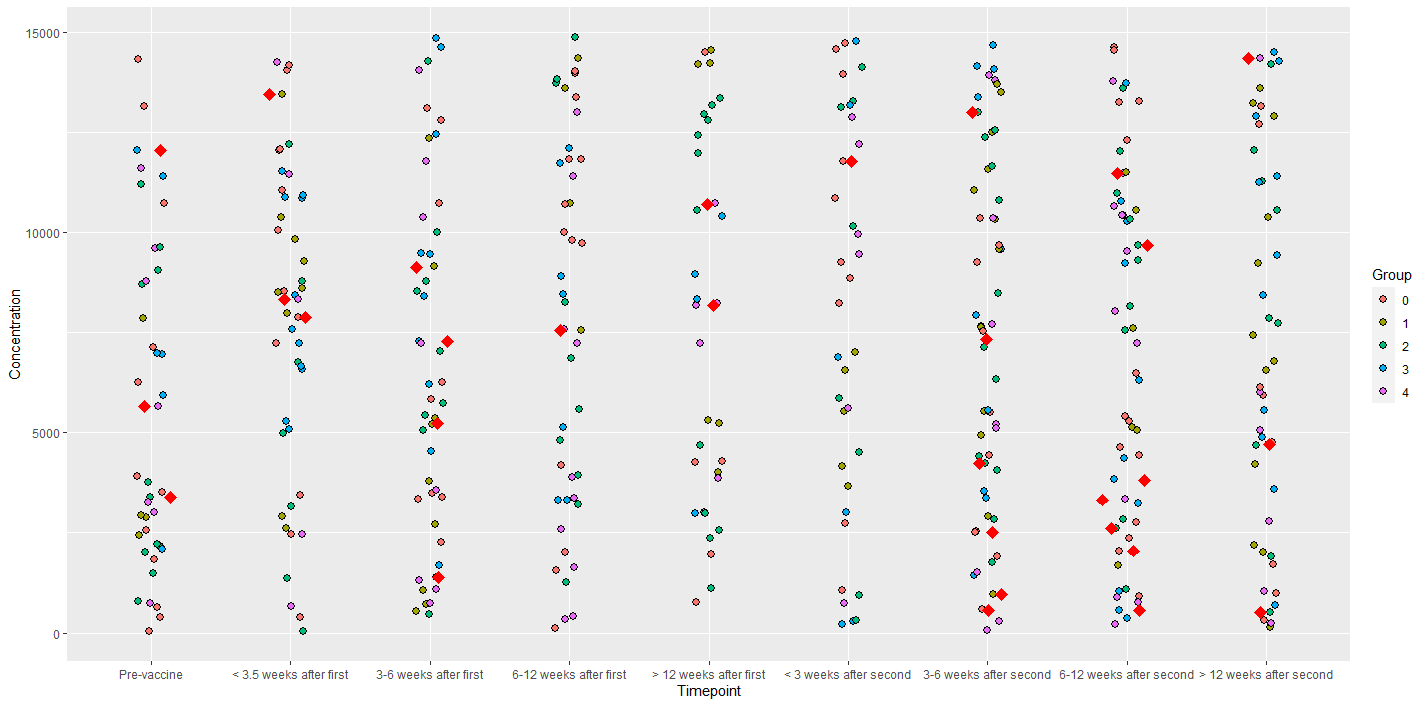
Now let's prepare the graph. The data you want to highlight should be redrawn. In my case, this is data for Sample divisible by 13.
df %>% ggplot(aes(Timepoint, Concentration, fill=Group))
geom_jitter(position = position_jitter(width = 0.1),pch = 21, size = 2.5)
geom_point(data = df %>% filter(Sample %% 13==0),
position = position_jitter(width = 0.2), pch = 23, size = 3,
fill="red", color = "red")
Note that I applied the data filter data = df %>% filter(Sample %% 13 == 0). You make your own filter adapted to the data you have.
Finally, one more thing. I completely don't understand why you are using geom_jitter and setting position_jitter(width = 0.0001). This is completely pointless. geom_jitter is just for making the data a bit spread out so that they don't overlap. However, when you set width = 0.0001, it's like you don't use jitter at all.
CodePudding user response:
I'm going to start by saying using red as a colour to mean one thing, while already using colour to mean something else is confusing.
You could plot a red circle round the point of interest. Or add an arrow.
As suggested gghighlight may have an option for you. Or it may not.
Without wanting to redraw your whole plots for my personal preference... ...can I suggest geom_beoswarm() from ggbeeswarm may may your plots more legible in terms of understanding the data distribution.
OK So now to address the underlying question. Always tricky when we don't have a sample of your data
require(ggpolot)
require(tidyverse)
seed(42)
someData <- tibble(
Timepoint = as.factor(rep(seq(0,8),10)),
Concentration = sample(1:100000, 90, replace=F),
Group = rep(seq(0,4), 18 )
) %>%
mutate( Sample = paste0("V",ceiling(row_number()/9)))
someData %>%
mutate(Group = as.factor(Group)) %>%
mutate(Timepoint = fct_recode(Timepoint, `Pre-vaccine` = "0",
"< 3.5 weeks after first" = "1",
"3-6 weeks after first" = "2",
"6-12 weeks after first" ="3",
"> 12 weeks after first" = "4",
"< 3 weeks after second" = "5",
"3-6 weeks after second" = "6",
"6-12 weeks after second" ="7" ,
"> 12 weeks after second" = "8")) -> someData
# You have defined some constants that aren't explained
pal <- c("V1" = "red", "0"= "Purple", "1" = "Blue", "2" = "Green", "3" = "Yellow", "4" = "Black", "5"="Pink")
# I've simply omitted breaks and minor_breaks from your code below
This is simply your graph working with the sample data above
someData %>%
ggplot(aes(Timepoint, Concentration))
geom_jitter(position = position_jitter(width = 0.0001),
aes(fill = ifelse(str_detect(Sample, "V1"), Sample, Group)), # Here is where I specify the colour fill of data points if they match the study ID, if not they are coloured by 'Group'
pch = 21,
size = 2.5)
scale_y_log10(labels = scales::comma,
limits = c(10,10000000),
#breaks = breaks,
#minor_breaks = minor_breaks
)
theme_classic()
labs(title = "Antibody levels",
x = "",
y = "Concentration (AU/ml)")
annotation_logticks(base = 10, sides = "l")
scale_fill_manual(values = pal)
theme(plot.title = element_text(hjust = 0.5),
axis.text.y = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1, face = "bold"),
legend.position = "none")
You can simply add a second geom_jitter over the first, the more recent line in the code sits above the other, and by only specifying a fill for the ones you want to highlight this achieves what you ask
someData %>%
ggplot(aes(Timepoint, Concentration))
geom_jitter(position = position_jitter(width = 0.0001),
aes(fill = Group),
pch = 21,
size = 2.5)
geom_jitter(position = position_jitter(width = 0.0001),
aes(fill = ifelse(str_detect(Sample, "V1"), "V1", NA)), # Here is where I specify the colour fill of data points if they match the study ID, if not they are coloured by 'Group'
pch = 21,
size = 2.5)
scale_y_log10(labels = scales::comma,
limits = c(10,10000000),
#breaks = breaks,
#minor_breaks = minor_breaks
)
theme_classic()
labs(title = "Antibody levels",
x = "",
y = "Concentration (AU/ml)")
annotation_logticks(base = 10, sides = "l")
scale_fill_manual(values = pal)
theme(plot.title = element_text(hjust = 0.5),
axis.text.y = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1, face = "bold"),
legend.position = "none")
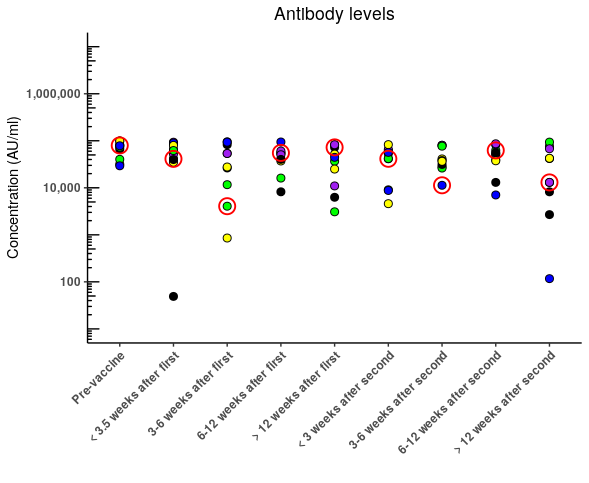
In my opinion - better - is to leave the fills as "Groups" as you intended but to highlight the key data points
# I've added a new palette that will highlight the sample of interest
pal2 <- c("V1" = "red", "V2"= NA, "V3" = NA, "V4" = NA, "V5" = NA,
"V6"=NA, "V7" = NA, "V8" = NA, "V9" = NA, "V10"= NA)
someData %>%
ggplot(aes(Timepoint, Concentration), warn)
geom_jitter(position = position_jitter(width = 0.0001),
aes(fill = Group),
pch = 21,
size = 2.5)
# You will get an error warning that some rows have missing values... thats becasue
# you only want to highlight some values
# If you need to - save the plot as an object using -> gg at the end
# and then suppressWarnings(print(gg))
geom_jitter(position = position_jitter(width = 0.0001),
aes( color=Sample, stroke = 1, fill = NA),
pch = 21,
size = 5)
scale_y_log10(labels = scales::comma,
limits = c(10,10000000),
#breaks = breaks,
#minor_breaks = minor_breaks
)
theme_classic()
labs(title = "Antibody levels",
x = "",
y = "Concentration (AU/ml)")
annotation_logticks(base = 10, sides = "l")
scale_fill_manual(values = pal)
scale_color_manual(values = pal2)
theme(plot.title = element_text(hjust = 0.5),
axis.text.y = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1, face = "bold"),
legend.position = "none")
For my own self indulgence
require(ggbeeswarm)
someData %>%
ggplot(aes(Timepoint, Concentration))
geom_beeswarm(
cex=1.75,
aes(fill = Group),
pch = 21,
size = 2.5)
scale_y_log10(labels = scales::comma,
limits = c(10,10000000),
#breaks = breaks,
#minor_breaks = minor_breaks
)
geom_beeswarm(
cex=1.75,
aes( color=Sample, stroke = 1, fill = NA),
pch = 21,
size = 5
)
theme_classic()
labs(title = "Antibody levels",
x = "",
y = "Concentration (AU/ml)")
annotation_logticks(base = 10, sides = "l")
scale_fill_manual(values = pal)
scale_color_manual(values = pal2)
theme(plot.title = element_text(hjust = 0.5),
axis.text.y = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1, face = "bold"),
legend.position = "none")