I'm running multiple pairwise comparisons on grouped factors and I would like to represent the resulting p-values with a heatmap.
This is a small example of what my dataset looks like, but in reality, it is much more complicated. My factors are site (2 levels) and organisms (3 levels).
site = c("SITE1","SITE1","SITE1","SITE1","SITE1","SITE1",
"SITE1","SITE1","SITE1","SITE2","SITE2","SITE2",
"SITE2","SITE2","SITE2","SITE2","SITE2","SITE2")
organism = c("Insects","Insects","Insects","Mammals","Mammals",
"Mammals","Reptiles","Reptiles","Reptiles","Insects",
"Insects","Insects","Mammals","Mammals","Mammals",
"Reptiles","Reptiles","Reptiles")
variable = c(5,6,7,12,13,14,1,2,3,7,8,9,22,24,25,11,12,14)
data = data.frame(site, organism, variable)
head(data)
site organism variable
1 SITE1 Insects 5
2 SITE1 Insects 6
3 SITE1 Insects 7
4 SITE1 Mammals 12
5 SITE1 Mammals 13
6 SITE1 Mammals 14
And this is how I'm computing the pairwise tests between pairs of organisms at each individual site:
data %>%
group_by(site) %>%
t_test(variable ~ organism)
site .y. group1 group2 n1 n2 statistic df p p.adj p.adj.signif
1 SITE1 variable Insects Mammals 3 3 -8.573214 4.000000 0.001000 0.002000 **
2 SITE1 variable Insects Reptiles 3 3 4.898979 4.000000 0.008000 0.008000 **
3 SITE1 variable Mammals Reptiles 3 3 13.472194 4.000000 0.000176 0.000528 ***
4 SITE2 variable Insects Mammals 3 3 -14.862705 3.448276 0.000300 0.000900 ***
5 SITE2 variable Insects Reptiles 3 3 -4.110961 3.448276 0.020000 0.020000 *
6 SITE2 variable Mammals Reptiles 3 3 9.086882 4.000000 0.000813 0.002000 **
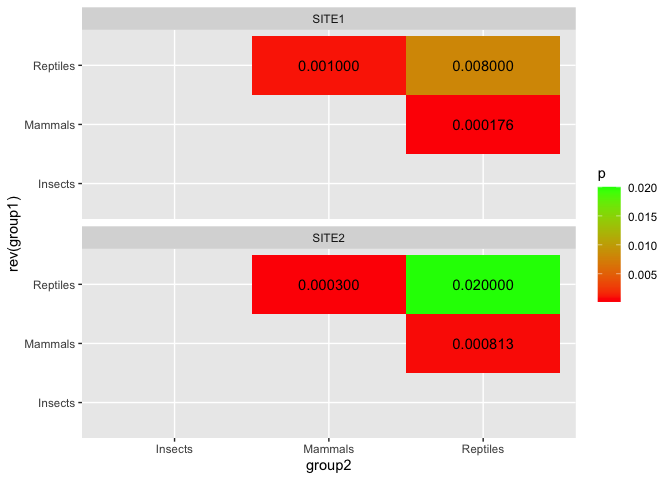
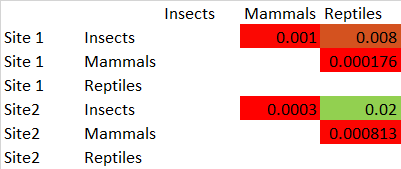
I would like to organise my data in a way that I can make a heatmap that would look similar to this:
I think the function pwpm from package emmeans does something similar, but it only works for emmeans. I was not able to find anything else.
Furthermore, it would be great if could also subdivide the p.values in p<0.05, p<0.01, p<0.001 for another heatmap so the significance is easier to see.
Could someone help me with this? I've been looking for hours without finding a way to do it.
CodePudding user response:
One option to achieve your desired result would be via ggplot2 and facetting. As your data is already in a tidy data format a basic heat map could be achieved via geom_tile facet_wrap. The rest is styling, like adding the p-values via geom_text, setting the colors via scale_fill_gradient or some additional data wrangling using e.g. tidyr::complete to add the "missing" group categories.
library(dplyr)
library(rstatix)
library(ggplot2)
d <- data %>%
group_by(site) %>%
t_test(variable ~ organism) %>%
tidyr::complete(group1 = unique(data$organism), group2 = unique(data$organism), site = unique(data$site))
ggplot(d, aes(group2, rev(group1), fill = p))
geom_tile()
geom_text(aes(label = scales::number(p, accuracy = 1e-6)))
scale_fill_gradient(low = "red", high = "green", na.value = NA)
facet_wrap(~site, ncol = 1)
#> Warning: Removed 12 rows containing missing values (geom_text).