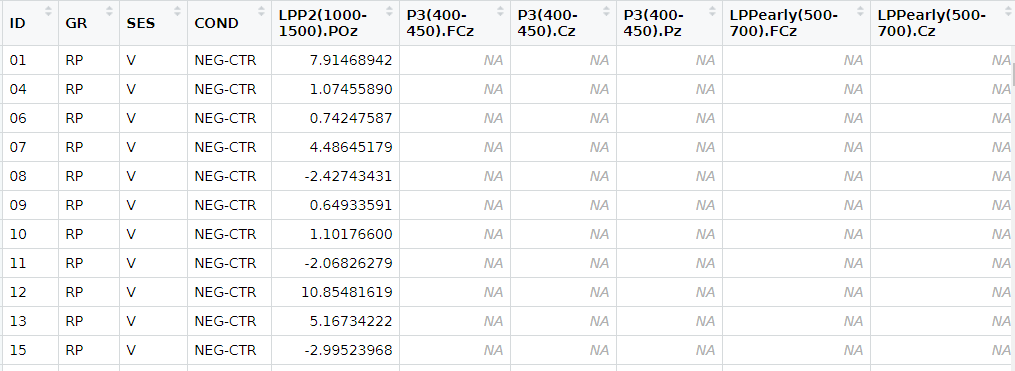
These are the datasets I'm working on.
Since the P-ones have the following structure
# A tibble: 25 x 5
ID GR SES COND `LPP2(1000-1500).POz`
<chr> <chr> <chr> <chr> <dbl>
1 01 RP V NEG-CTR 7.91
2 04 RP V NEG-CTR 1.07
3 06 RP V NEG-CTR 0.742
4 07 RP V NEG-CTR 4.49
5 08 RP V NEG-CTR -2.43
6 09 RP V NEG-CTR 0.649
7 10 RP V NEG-CTR 1.10
8 11 RP V NEG-CTR -2.07
9 12 RP V NEG-CTR 10.9
10 13 RP V NEG-CTR 5.17
While the V-ones have this following ones, with a more extended number of coulmns:
> `RP-V-NEG-CTR`
# A tibble: 25 x 16
ID GR SES COND `P3(400-450).FCz` `P3(400-450).Cz` `P3(400-450).Pz` `LPPearly(500-700)~
<chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 01 RP V NEG-CTR -11.6 -5.17 11.9 -11.8
2 04 RP V NEG-CTR -0.314 2.23 15.1 -4.02
3 06 RP V NEG-CTR -0.214 -1.30 3.14 4.47
4 07 RP V NEG-CTR -2.83 2.19 13.7 -0.884
5 08 RP V NEG-CTR 4.24 2.45 7.48 1.15
6 09 RP V NEG-CTR 9.57 13.7 23.4 13.3
7 10 RP V NEG-CTR -6.13 -4.13 6.27 0.0229
8 11 RP V NEG-CTR 0.529 5.43 13.0 1.90
9 12 RP V NEG-CTR -7.74 -2.41 15.8 -8.08
10 13 RP V NEG-CTR 1.27 3.48 9.94 1.75
# ... with 15 more rows, and 8 more variables: LPPearly(500-700).Cz <dbl>,
# LPPearly(500-700).Pz <dbl>, LPP1(500-1000).FCz <dbl>, LPP1(500-1000).Cz <dbl>,
# LPP1(500-1000).Pz <dbl>, LPP2(1000-1500).FCz <dbl>, LPP2(1000-1500).Cz <dbl>,
# LPP2(1000-1500).Pz <dbl>
>
I've used the followinf procedure for importing and merging them together:
files <- list.files(pattern = "\\.xls")
for (i in 1:length(files)) {
assign(gsub("\\.xls", "", files[i]), readxl::read_xls(files[i]))
}
data <- `RP-V-NEG-CTR` %>%
add_row(`RP-V-NEG-NOC`) %>%
add_row(`RP-V-NEU-NOC`)%>%
arrange(ID)
data1 <- `RP-POz-NEG-CTR` %>%
add_row(`RP-POz-NEG-NOC`) %>%
add_row(`RP-POz-NEU-NOC`)%>%
arrange(ID)
data <- merge(data, data1, by = c('ID', 'GR', 'SES', 'COND'))
Since trying by this following code I've obtain a non-matched merged dataset:
filenames <- list.files(pattern = '^RP.*\\.xls$')
> data <- purrr::map_df(filenames, readxl::read_excel)
Like this:
In the case I would ike to import already merged in a proper way, what am I suppsed to do/adjust?
Thanks in advance
CodePudding user response:
Maybe we need map2
library(dplyr)
library(purrr)
library(stringr)
filesv <- list.files(pattern = 'RP-V-.*\\.xls', full.names = TRUE)
filesp <- list.files(pattern = 'RP-P-.*\\.xls', full.names = TRUE)
nm1 <- str_c(str_remove(basename(filesv), "\\.xls"),
str_remove(basename(filesp), "\\.xls"), sep="_")
out <- map2(filesv, filesp, ~ {
vdat <- readxl::read_excel(.x)
pdat <- readxl::read_excel(.y)
inner_join(vdat, pdat, by = c('ID', 'GR', 'SES', 'COND'))
}) %>%
setNames(nm1) %>%
bind_rows(.id = 'grp')
Or if we don't need the 'grp' column
out <- map2_dfr(filesv, filesp, ~ {
vdat <- readxl::read_excel(.x)
pdat <- readxl::read_excel(.y)
inner_join(vdat, pdat, by = c('ID', 'GR', 'SES', 'COND'))
})