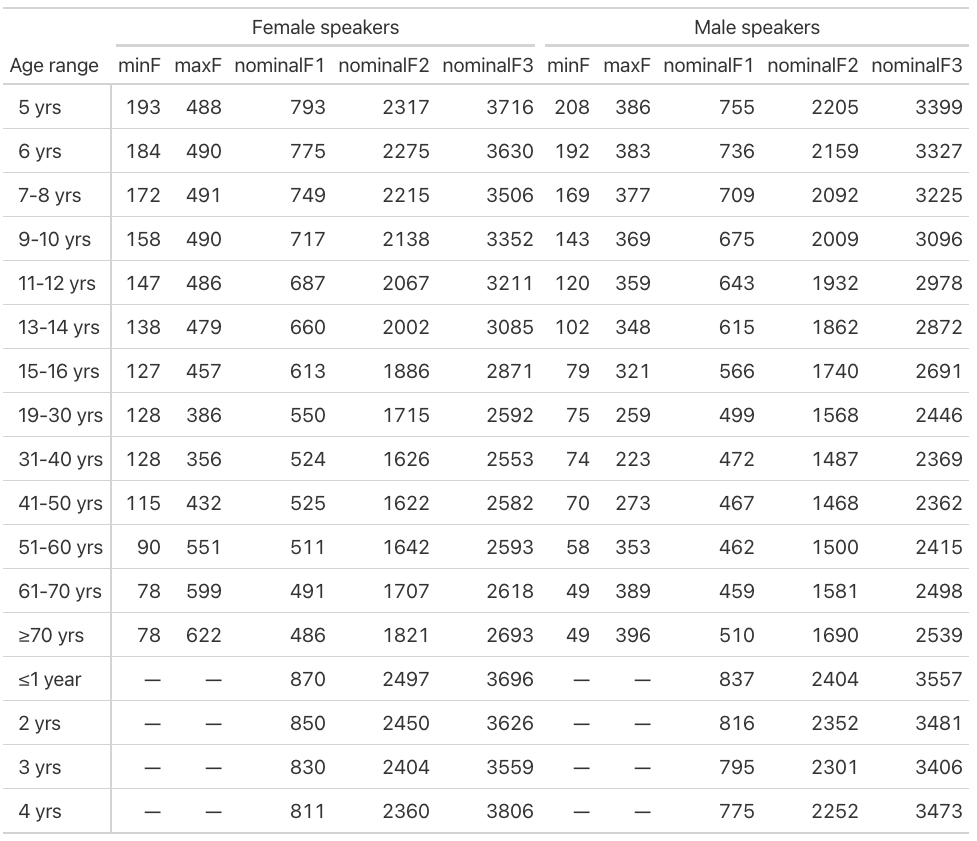
I have a silly problem in making a table using the otherwise excellent gt package.
I have this table:
currTab <- structure(list(`Age range` = structure(c(5L, 6L, 7L, 8L, 9L,
10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 1L, 2L, 3L, 4L), .Label = c(" ≤1 year",
"2 yrs", "3 yrs", "4 yrs", "5 yrs", "6 yrs", "7-8 yrs", "9-10 yrs",
"11-12 yrs", "13-14 yrs", "15-16 yrs", "19-30 yrs", "31-40 yrs",
"41-50 yrs", "51-60 yrs", "61-70 yrs", " ≥70 yrs"), class = c("ordered",
"factor")), minF_Female = c(192.513259215355, 183.831034165788,
171.923523427238, 158.002976410505, 146.564781760376, 137.789704831087,
127.370737204799, 128.398075852923, 127.830665246427, 114.61707107954,
90.3633466641061, 78.010313243078, 78.1870193570691, NA, NA,
NA, NA), minF_Male = c(207.855719860607, 191.535711870608, 169.079769539432,
142.555089696259, 120.190679816665, 102.112304160514, 79.4522096443978,
74.5501293799995, 74.0560562513096, 69.8375905638724, 57.7437980326593,
49.4092475281028, 48.9416497200912, NA, NA, NA, NA), maxF_Female = c(488.066672846171,
489.629983633728, 490.630420606523, 489.6953109154, 485.997969341596,
479.493366060129, 457.131832057242, 386.052841894336, 356.20654778542,
432.373754791911, 550.984958319451, 599.030148485128, 622.392016651455,
NA, NA, NA, NA), maxF_Male = c(386.491941716704, 383.013074412721,
377.262604560802, 368.664103740006, 358.88619036232, 347.858049187743,
321.425630395873, 258.610024319393, 223.40438098279, 272.562362517114,
352.663065356397, 389.139726222453, 396.387425129329, NA, NA,
NA, NA), nominalF1_Female = c(792.606585070623, 774.67333252336,
748.978655452818, 716.766904166985, 687.067342631714, 659.942877422917,
613.464127049404, 549.839814945283, 524.453021680478, 524.604252664451,
511.016250159981, 490.803284150502, 486.151004978768, 870.189435502058,
849.931906280897, 830.243701660919, 811.132651353652), nominalF1_Male = c(754.927353762102,
735.91406180303, 708.672474530183, 674.554785316654, 643.185788473706,
614.682090933963, 565.988043403975, 499.441794119581, 471.850496193325,
467.279224258147, 461.886047442941, 458.911042417315, 509.566754276534,
837.039982744214, 815.632443409745, 794.801313106926, 774.561360377223
), nominalF2_Female = c(2316.79654051926, 2274.86292176996, 2214.51473531373,
2138.33574040433, 2067.34402899125, 2001.61879507854, 1885.76397373785,
1714.61348292451, 1625.93585114191, 1622.14234117408, 1641.9648727809,
1707.39999135551, 1820.70094063161, 2497.1503947177, 2450.18163132232,
2404.46219881265, 2359.99840070589), nominalF2_Male = c(2204.72154293521,
2158.60270220094, 2092.40242942714, 2009.22489043384, 1932.3313895761,
1861.92275643787, 1740.32902601832, 1567.84624090872, 1487.1049441955,
1467.78392743302, 1499.7012515019, 1581.48607357091, 1689.73201386241,
2403.51904879437, 2351.72182188825, 2301.30633827736, 2252.29783346018
), nominalF3_Female = c(3716.13980758452, 3629.61667155808, 3506.02757238294,
3351.95172828646, 3211.26448866239, 3084.53931579307, 2871.44540359288,
2591.76336829096, 2553.30039539724, 2581.55257096308, 2593.29167872634,
2617.84457067156, 2692.78060751014, 3695.7825841822, 3626.26881435704,
3558.60405424272, 3805.66484270251), nominalF3_Male = c(3398.97976277625,
3327.2795690819, 3224.61918529811, 3096.22641948377, 2978.49201056792,
2871.90468878882, 2690.5686257907, 2445.67764464339, 2369.21560078373,
2361.71802712342, 2414.55734269414, 2497.82856943456, 2539.1115058239,
3557.20819221567, 3480.54829639461, 3405.93338065049, 3473.04043452418
)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA,
-17L))
in which the Age range column is a factor:
> levels(currTab$`Age range`)
[1] " ≤1 year" "2 yrs" "3 yrs" "4 yrs" "5 yrs" "6 yrs" "7-8 yrs" "9-10 yrs" "11-12 yrs" "13-14 yrs" "15-16 yrs"
[12] "19-30 yrs" "31-40 yrs" "41-50 yrs" "51-60 yrs" "61-70 yrs" " ≥70 yrs"
and apply this formating code:
currTab %>%
gt(rowname_col = "Age range") %>%
tab_stubhead(label="Age range") %>%
tab_spanner(
label = "Female speakers",
columns = c(minF_Female,maxF_Female,nominalF1_Female, nominalF2_Female,nominalF3_Female)
) %>%
tab_spanner(
label = "Male speakers",
columns = c(minF_Male,maxF_Male,nominalF1_Male, nominalF2_Male,nominalF3_Male)
) %>%
cols_label(
minF_Female = "minF",
maxF_Female = "maxF",
nominalF1_Female = "nominalF1",
nominalF2_Female = "nominalF2",
nominalF3_Female = "nominalF3" ,
minF_Male = "minF",
maxF_Male = "maxF",
nominalF1_Male = "nominalF1",
nominalF2_Male = "nominalF2",
nominalF3_Male = "nominalF3"
) %>%
fmt_integer(
columns = minF_Female:nominalF3_Male,
use_seps=FALSE
) %>%
fmt_missing(everything(),missing_text="---")
and get this table:
As you can imagine, I would prefer the Age range column to follow the factor levels, and not place the first ones with empty columns at the end like that.
Any ideas?
Fredrik
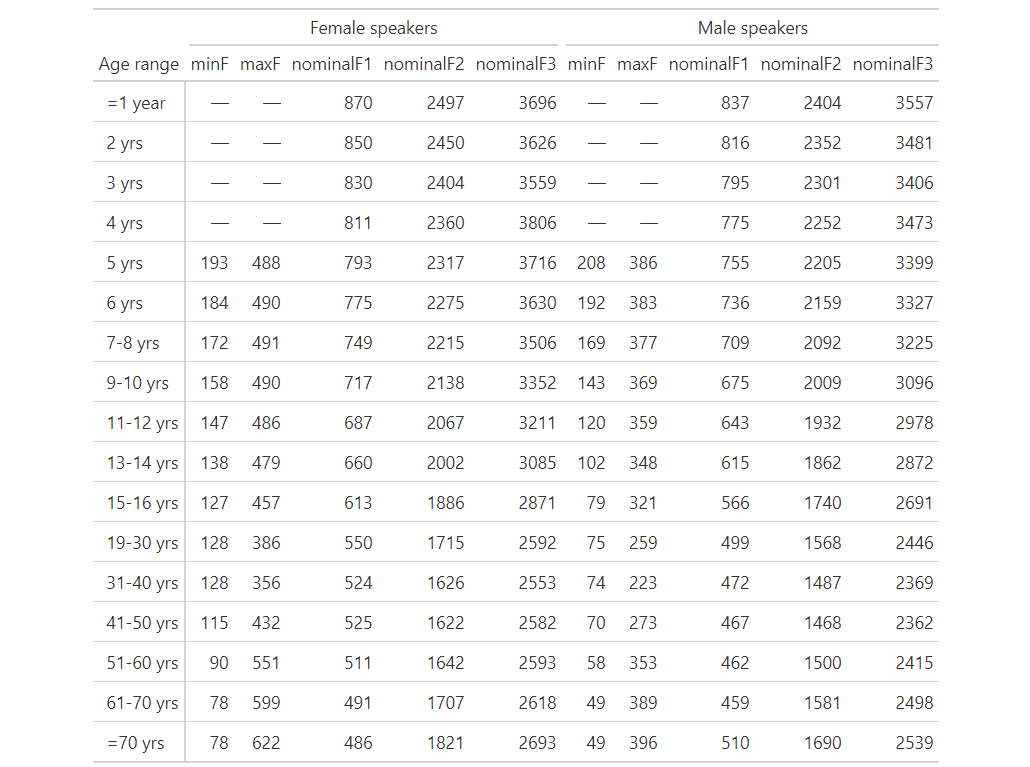
CodePudding user response:
Order the dataframe beforehand:
base R
currTab[order(currTab$`Age range`),]
dplyr
currTab %>%
arrange(`Age range`)
output
currTab %>%
arrange(`Age range`) %>%
gt(rowname_col = "Age range") %>%
tab_stubhead(label="Age range") %>%
tab_spanner(
label = "Female speakers",
columns = c(minF_Female,maxF_Female,nominalF1_Female, nominalF2_Female,nominalF3_Female)
) %>%
tab_spanner(
label = "Male speakers",
columns = c(minF_Male,maxF_Male,nominalF1_Male, nominalF2_Male,nominalF3_Male)
) %>%
cols_label(
minF_Female = "minF",
maxF_Female = "maxF",
nominalF1_Female = "nominalF1",
nominalF2_Female = "nominalF2",
nominalF3_Female = "nominalF3" ,
minF_Male = "minF",
maxF_Male = "maxF",
nominalF1_Male = "nominalF1",
nominalF2_Male = "nominalF2",
nominalF3_Male = "nominalF3"
) %>%
fmt_integer(
columns = minF_Female:nominalF3_Male,
use_seps=FALSE
) %>%
fmt_missing(everything(),missing_text="---")