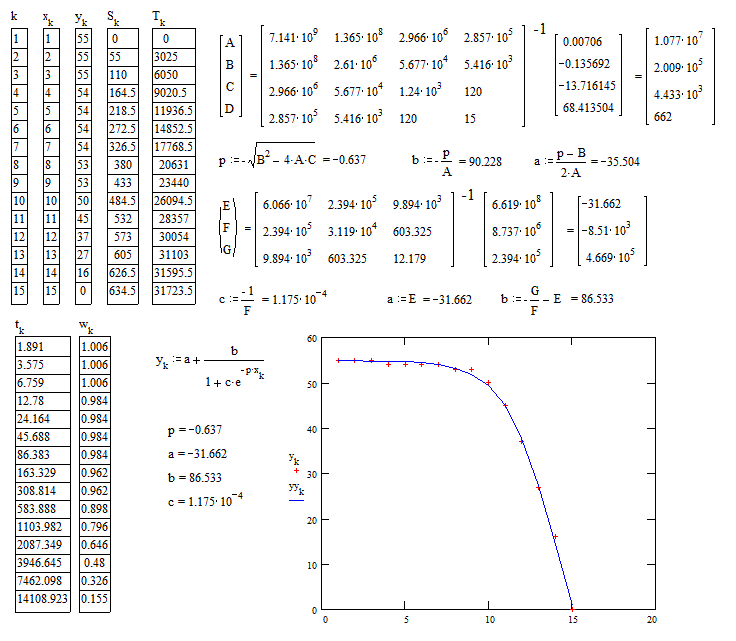
I am unable to properly fit a logarithmic and exponential decay curve to my experimental data points, where it is as if the suggested curve fits do not resemble the pattern in my data not even remotely.
I have the following example data:
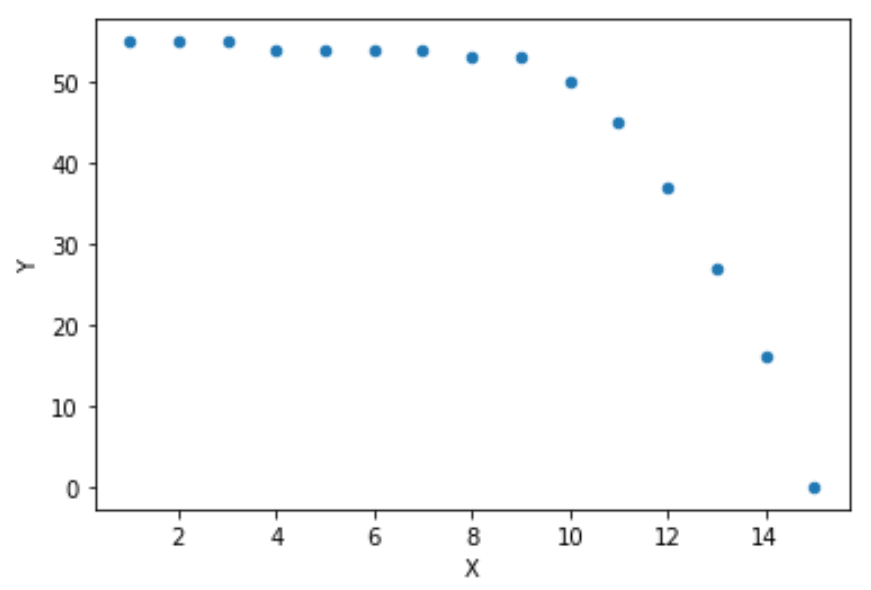
data = {'X':[1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15
],
'Y':[55, 55, 55, 54, 54, 54, 54, 53, 53, 50, 45, 37, 27, 16, 0
]}
df = pd.DataFrame(data)
df = pd.DataFrame(data,columns=['X','Y'])
df.plot(x ='X', y='Y', kind = 'scatter')
plt.show()
I then try fitting an exponential decay and logarithmic decay curve to these data points using this code and outputting the root mean square error for each curve:
# load the dataset
data = df.values
# choose the input and output variables
x, y = data[:, 0], data[:, 1]
def func1(x, a, b, c):
return a*exp(b*x) c
def func2(x, a, b):
return a * np.log(x) b
params, _ = curve_fit(func1, x, y)
a, b, c = params[0], params[1], params[2]
yfit1 = a*exp(x*b) c
rmse = np.sqrt(np.mean((yfit1 - y) ** 2))
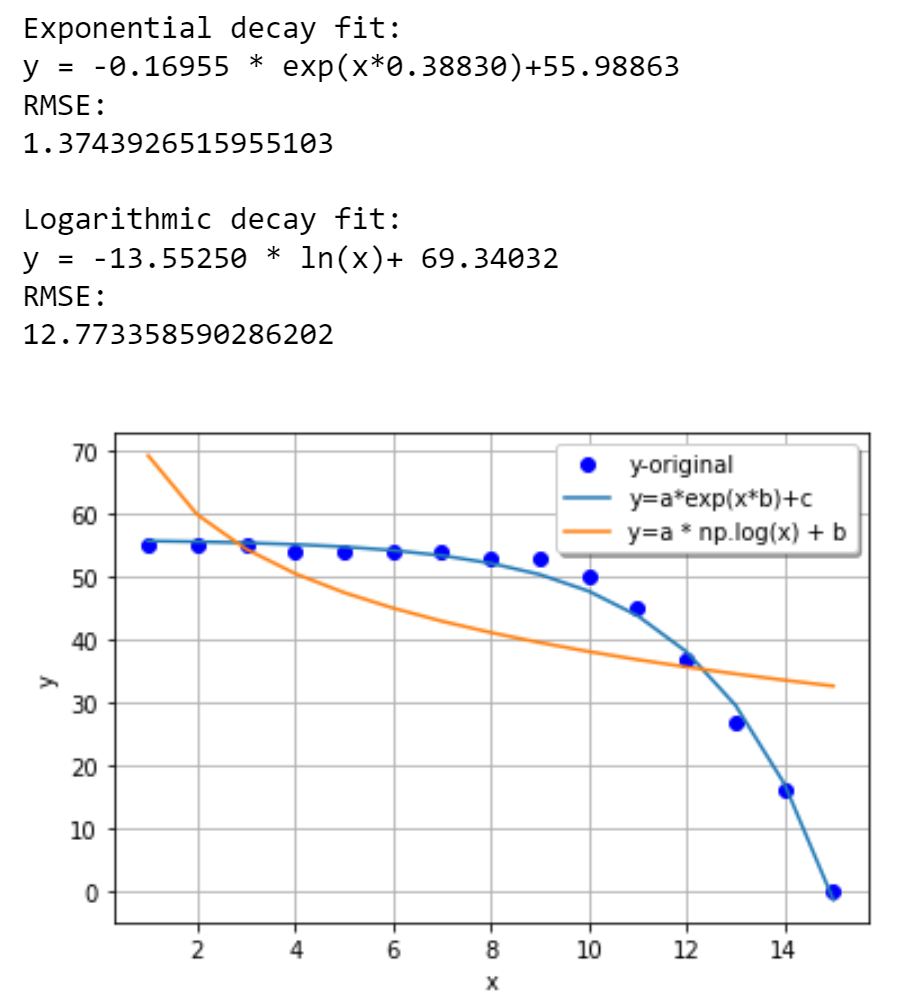
print('Exponential decay fit:')
print('y = %.5f * exp(x*%.5f) %.5f' % (a, b, c))
print('RMSE:')
print(rmse)
print('')
params, _ = curve_fit(func2, x, y)
a, b = params[0], params[1]
yfit2 = a * np.log(x) b
rmse = np.sqrt(np.mean((yfit2 - y) ** 2))
print('Logarithmic decay fit:')
print('y = %.5f * ln(x) %.5f' % (a, b))
print('RMSE:')
print(rmse)
print('')
plt.plot(x, y, 'bo', label="y-original")
plt.plot(x, yfit1, label="y=a*exp(x*b) c")
plt.plot(x, yfit2, label="y=a * np.log(x) b")
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='best', fancybox=True, shadow=True)
plt.grid(True)
plt.show()
I then try to using my experimental data, trying these new data points:
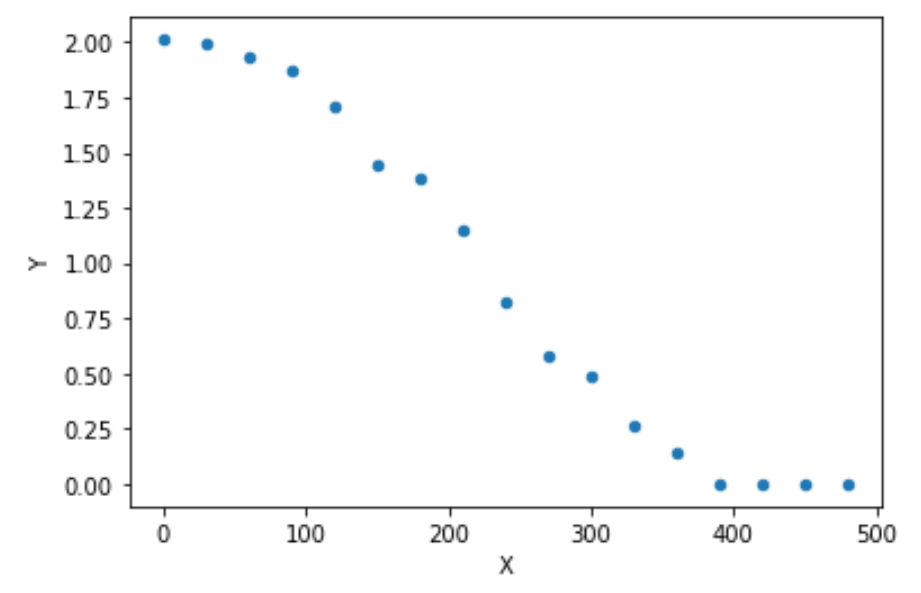
data = {'X':[0, 30, 60, 90, 120, 150, 180, 210, 240, 270, 300, 330, 360, 390, 420, 450, 480
],
'Y':[2.011399983,1.994139959,1.932761226,1.866343728,1.709889128,1.442674671,1.380548494,1.145193671,0.820646118,
0.582299012, 0.488162766, 0.264390575, 0.139457758, 0, 0, 0, 0
]}
df = pd.DataFrame(data)
df = pd.DataFrame(data,columns=['X','Y'])
df.plot(x ='X', y='Y', kind = 'scatter')
plt.show()
I then try using the previous code to fit an exponential decay curve and a logarithmic decay curve to these new data points with this:
import pandas as pd
import numpy as np
from numpy import array, exp
from scipy.optimize import curve_fit
import matplotlib.pyplot as plt
# load the dataset
data = df.values
# choose the input and output variables
x, y = data[:, 0], data[:, 1]
def func1(x, a, b, c):
return a*exp(b*x) c
def func2(x, a, b):
return a * np.log(x) b
params, _ = curve_fit(func1, x, y)
a, b, c = params[0], params[1], params[2]
yfit1 = a*exp(x*b) c
rmse = np.sqrt(np.mean((yfit1 - y) ** 2))
print('Exponential decay fit:')
print('y = %.5f * exp(x*%.5f) %.5f' % (a, b, c))
print('RMSE:')
print(rmse)
print('')
params, _ = curve_fit(func2, x, y)
a, b = params[0], params[1]
yfit2 = a * np.log(x) b
rmse = np.sqrt(np.mean((yfit2 - y) ** 2))
print('Logarithmic decay fit:')
print('y = %.5f * ln(x) %.5f' % (a, b))
print('RMSE:')
print(rmse)
print('')
plt.plot(x, y, 'bo', label="y-original")
plt.plot(x, yfit1, label="y=a*exp(x*b) c")
plt.plot(x, yfit2, label="y=a * np.log(x) b")
plt.xlabel('x')
plt.ylabel('y')
plt.legend(loc='best', fancybox=True, shadow=True)
plt.grid(True)
plt.show()
And I receive this output which looks totally wrong:
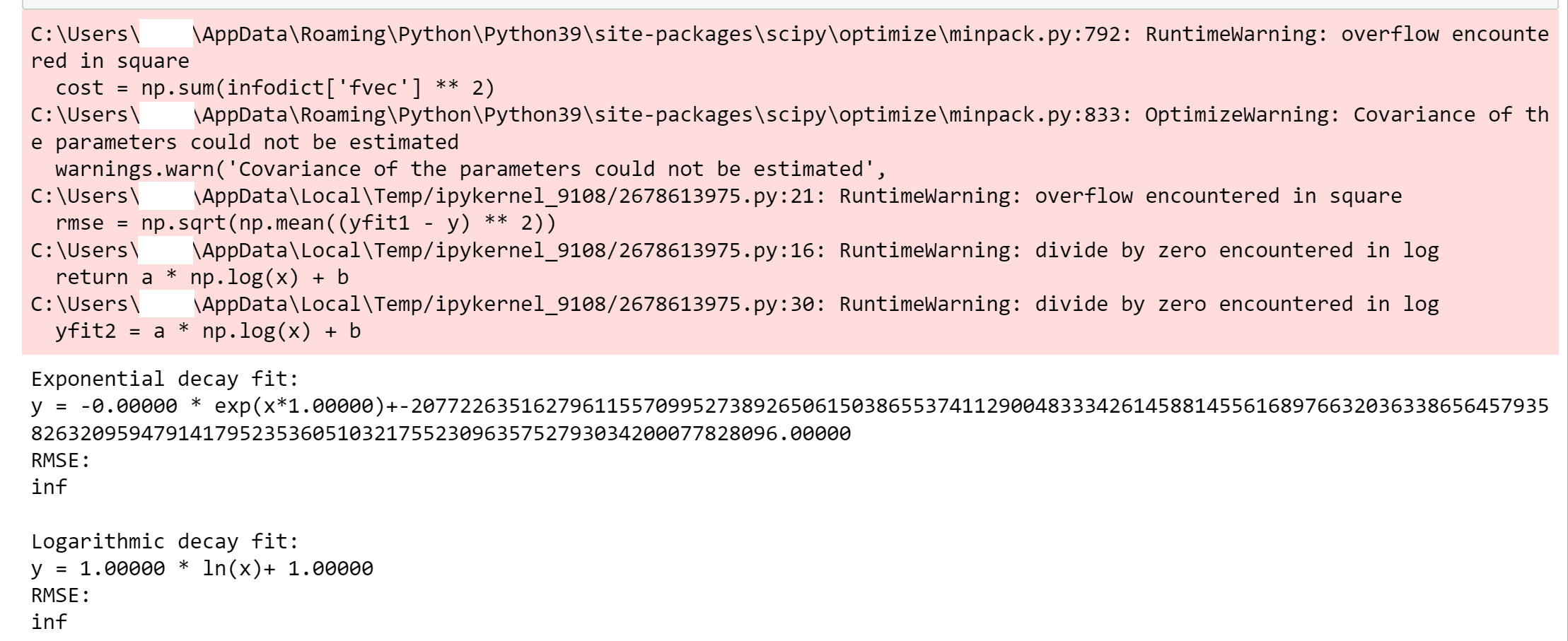
And then I receive this plotted output which looks very far off from my experimental data points:
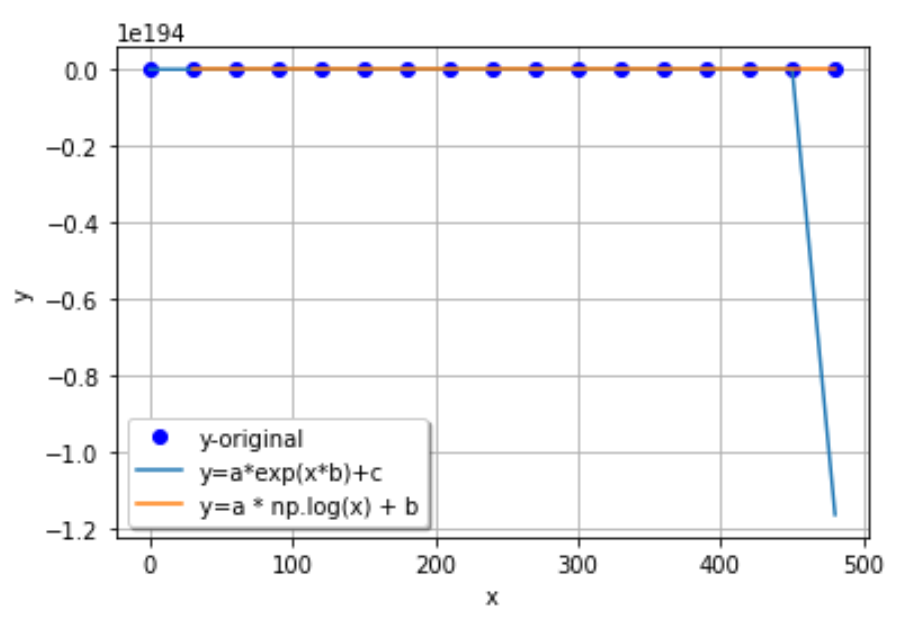
I do not understand why my first curve fitting attempt worked so well and smoothly, while my second attempt seems to have turned into a huge incoherent mess that just broke the curve_fit function. I do not understand why I see the graph going into the negative y-axis if I do not have any negative y-axis values in my experimental data. I am confused because I can clearly see my experimental data plotted fine as just points, so I am not sure what is so wrong about it that I cannot simply fit my curves to the points. How can I address my code so that I can properly use curve_fit() to fit an exponential decay curve and a logarithmic decay curve to my experimental data points?
CodePudding user response:
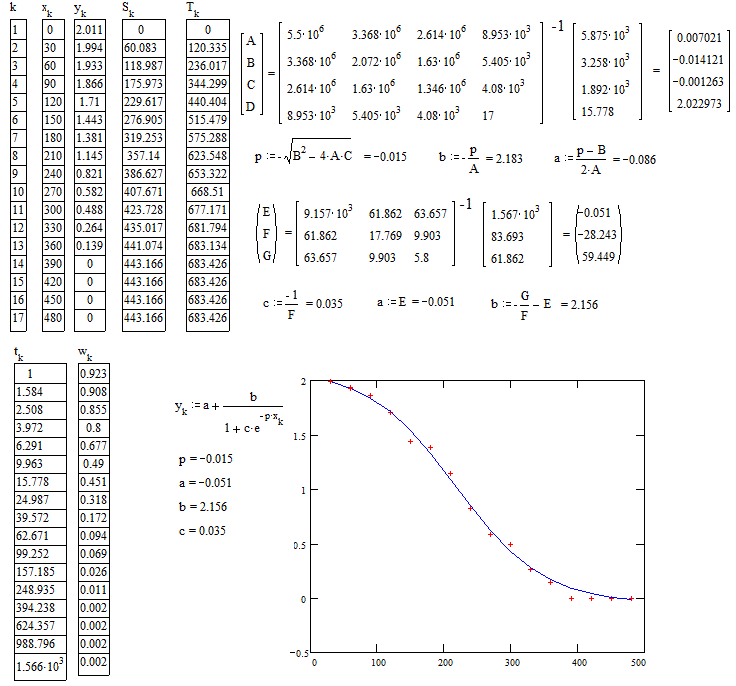
As already pointed out in comments the model seems on the logistic kind.
The main difficulty for fitting with the usual softwares is the choice of the initial values of the parameters to start the iterative calculus. A non conventional method which general principle is explained in 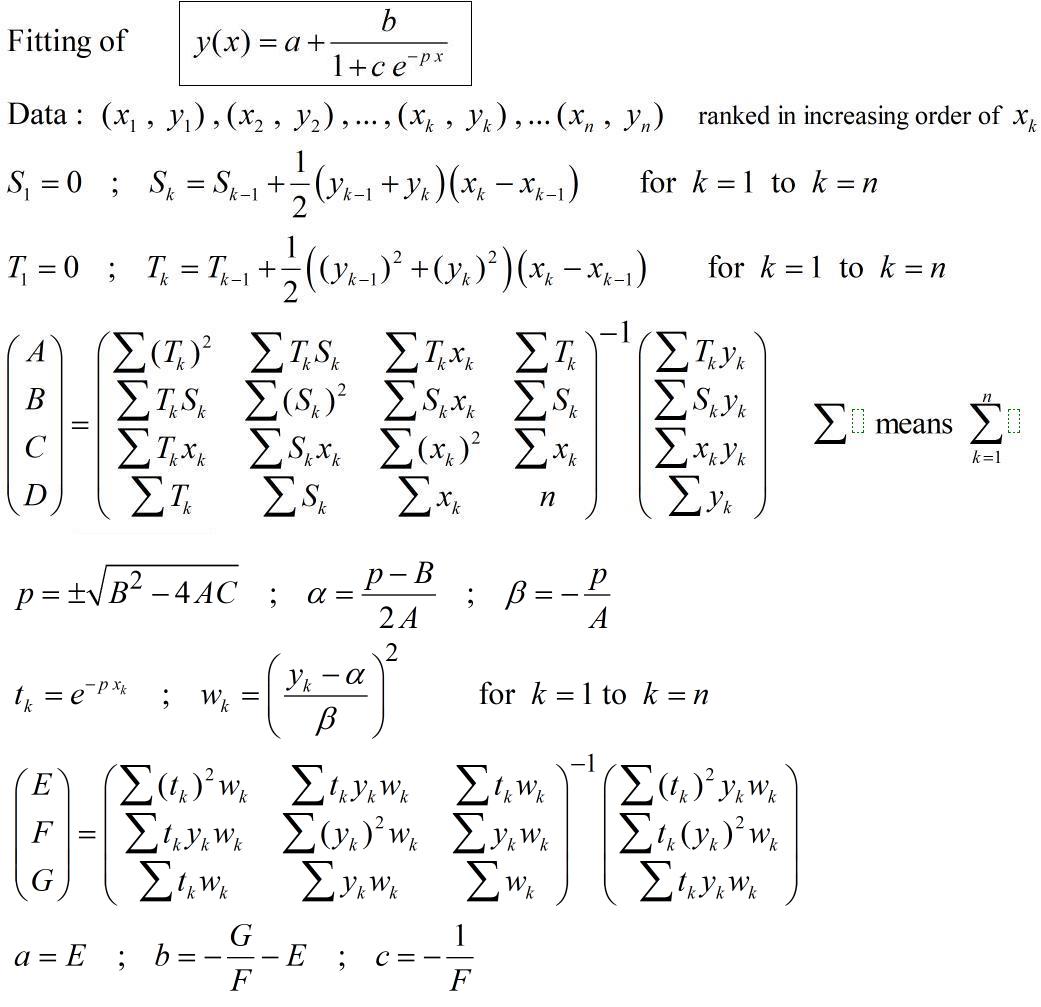
With your second data :
With your first data :
If you want a more accurate fit according to some specified criteria of fitting (MSE, MSRE, MAE, or other) you could take the above values of parameters as starting values in a non-linear regression software.