I am conducting a test with two factorial parameters (x, y) in an image processing program, which has produced a number of test images as pngs of the same dimensions. For example, in the mtcars dataset, they could represent one illustrative car image for each of the cyl/carb combinations.
I would like to import these images into R and plot them in a facet grid with the respective parameter values on the axes (e.g. cyl and carb).
What's the best way to a) import the images into a tibble/df, b) plotting them as per above?
(If necessary, I'd be happy to update the question with example code once I know what package to use).
CodePudding user response:
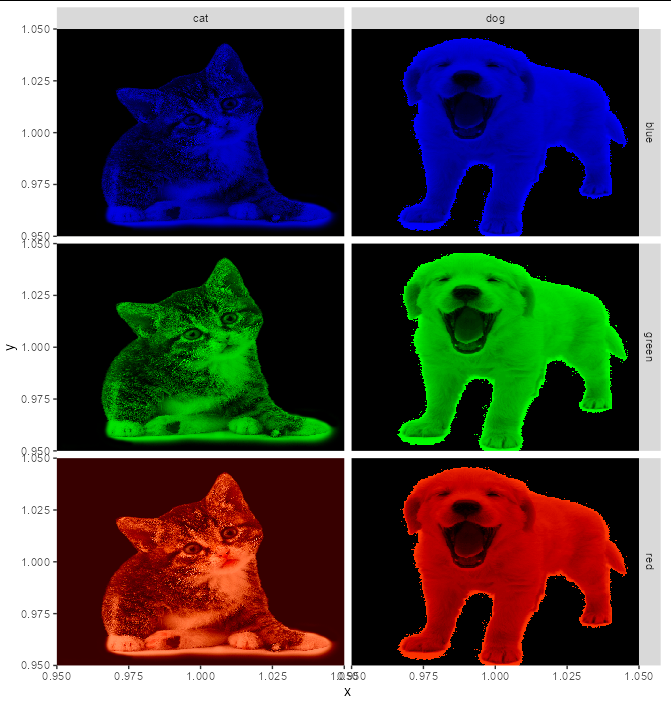
If you have the images as png, you don't actually need to convert them to data frames. You can use geom_image from ggimage, which only requires the file path. Suppose I have two categories - "animal" and "color", with an image representing the conjunction of each unique "color" and "animal".
I need only do:
df <- data.frame(animal = rep(c("cat", "dog"), each = 3),
color = rep(c("red", "green", "blue"), 2),
img = path.expand(
c("~/redcat.png", "~/greencat.png", "~/bluecat.png",
"~/reddog.png", "~/greendog.png", "~/bluedog.png")))
library(ggimage)
ggplot(df)
geom_image(aes(x = 1, y = 1, image = img), size = 1)
facet_grid(color~animal, scales = "free")