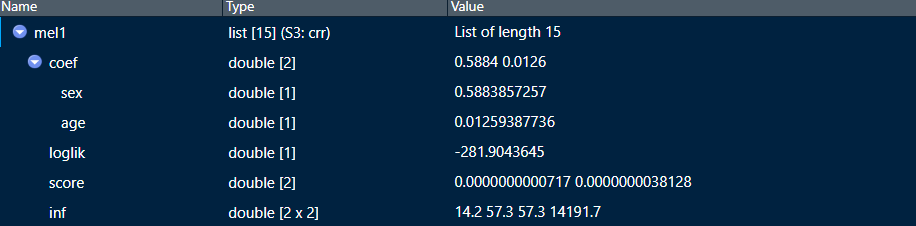
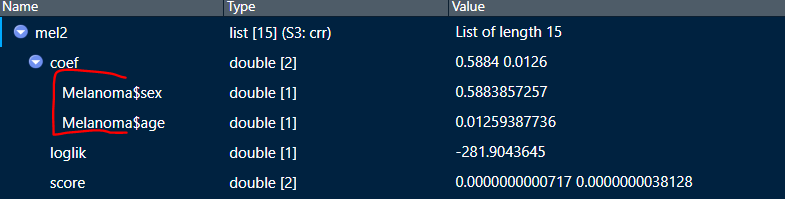
I am using the cmprsk package to create a series of regressions. In the real models I used, I specified my models in the same way that is shown in the example that produces mel2 below. My problem is, I want the Melanoma$ in front of the coefficients to go away, as happens if I had specified the model like in mel1. Is there a way to delete that data frame prefix out of the object without re-running it?
library(cmprsk)
data(Melanoma, package = "MASS")
head(Melanoma)
mel1 <- crr(ftime = Melanoma$time, fstatus = Melanoma$status, cov1 = Melanoma[, c("sex", "age")], cencode = 2)
covs2 <- model.matrix(~ Melanoma$sex Melanoma$age)[, -1]
mel2 <- crr(ftime = Melanoma$time, fstatus = Melanoma$status, cov1 = covs2, cencode = 2)
CodePudding user response:
You could use the data argument in model.matrix, and wrap the crr call in with(Melanoma, ...)
covs2 <- model.matrix(~ sex age, data = Melanoma)[, -1]
mel2 <- with(Melanoma, crr(ftime = time, fstatus = status,
cov1 = covs2, cencode = 2))
mel2$coef
#> sex age
#> 0.58838573 0.01259388
If you are stuck with existing models like this:
covs2 <- model.matrix(~ Melanoma$sex Melanoma$age)[, -1]
mel2 <- crr(ftime = Melanoma$time, fstatus = Melanoma$status,
cov1 = covs2, cencode = 2)
You could simply rename the coefficients like this
names(mel2$coef) <- c("sex", "age")
mel2
#> convergence: TRUE
#> coefficients:
#> sex age
#> 0.58840 0.01259
#> standard errors:
#> [1] 0.271800 0.009301
#> two-sided p-values:
#> sex age
#> 0.03 0.18