I have several csv files which have data of voltage over time and each csv files are approximately 7000 rows and the data looks like this:
Time(us) Voltage (V)
0 32.96554106
0.5 32.9149649
1 32.90484966
1.5 32.86438874
2 32.8542735
2.5 32.76323642
3 32.74300595
3.5 32.65196886
4 32.58116224
4.5 32.51035562
5 32.42943376
5.5 32.38897283
6 32.31816621
6.5 32.28782051
7 32.26759005
7.5 32.21701389
8 32.19678342
8.5 32.16643773
9 32.14620726
9.5 32.08551587
10 32.04505495
10.5 31.97424832
11 31.92367216
11.5 31.86298077
12 31.80228938
12.5 31.78205891
13 31.73148275
13.5 31.69102183
14 31.68090659
14.5 31.67079136
15 31.64044567
15.5 31.59998474
16 31.53929335
16.5 31.51906288
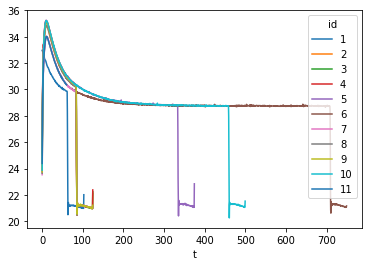
I read the csv file with pandas dataframe and after plotting a figure in matplotlib with data from one csv file, the figure looks like below.
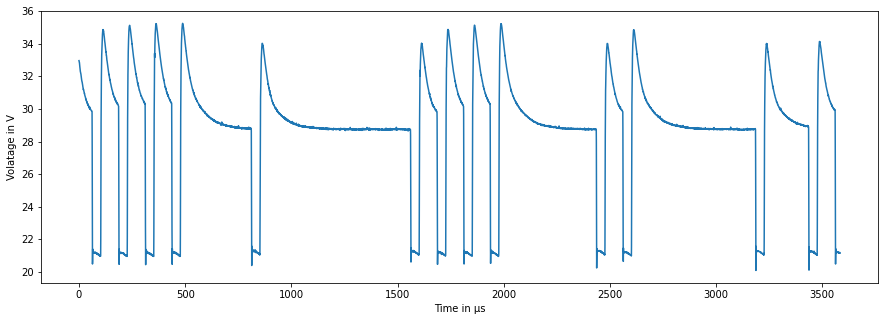
I would like to split every single square waveform/bit and store the corresponding voltage values for each bit separately. So the resulting voltage values of each bit would be stored in a row and should look like this:
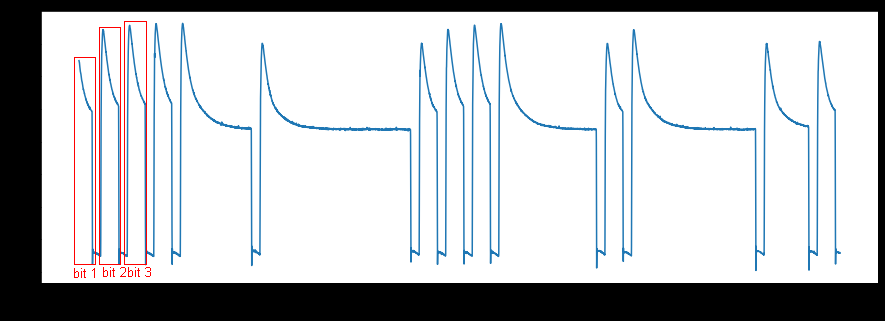
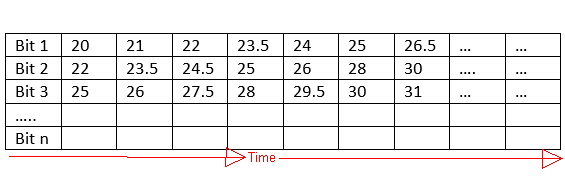
I don't have any idea how to do that. I guess I have to write a function where I have to assign a threshold value that, if the voltage values are going down for maybe 20 steps of time than capture all the values or if the voltage level is going up for 20 steps of time than capture all the voltage values. Could someone help?
CodePudding user response:
If you get the gradient of your Voltage (here using 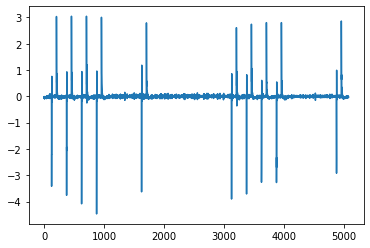
You can thus easily use a threshold (I tested with 2) to identify the peak starts. Then