I have fish stomach contents/diet data and I would like to get presence/absence information for each taxa in my df by month. Each observation (row) has information on the taxa absent (== 0) or present (== 1) in each fish's stomach. I have already transformed my original data to presence/absence values, however, I am not sure how to obtain a summary of what taxa was present or absent by month.
structure(list(id = c("607_6", "808_4", "801_3", "807_11", "801_16",
"724_13", "1030_40", "723_78", "701_4", "634_2", "1023_2", "1031_2",
"643_4", "606_3", "723_79", "801_4", "629_4", "642_10", "801_10",
"801_11", "1001_35", "616_4", "701_9", "627_2", "601_5"), Daphnia = c(0,
0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0), Byths = c(0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0,
1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1), Chiro.Pupae = c(0, 1, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0
), Empty = c(0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L), Chiro.Larvae = c(0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0), Amphipod = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0), Isopod = c(0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0),
Chironomidae = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Hemimysis = c(0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0), Copepoda = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0), Sphaeriidae = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0), Chiro.Adult = c(0, 0, 0, 0, 0, 0, 0, 1, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Trichopteran = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 1, 0, 0, 0), UID.Fish = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Chydoridae = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0), Cyclopoid = c(0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Fish.Eggs = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0), EggMass = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Dreissena = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0), Goby = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Eurycercidae = c(0,
0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0), Hirudinea = c(0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), totalnumPrey = c(0,
5, 0, 0, 102, 7, 220, 45, 0, 0, 0, 25, 116, 49, 119, 0, 7,
5, 0, 0, 0, 595, 105, 58, 20), MONTH = c(6L, 8L, 8L, 8L,
8L, 7L, 11L, 7L, 7L, 6L, 11L, 11L, 6L, 6L, 7L, 8L, 6L, 6L,
8L, 8L, 11L, 6L, 7L, 6L, 6L), empty = c("Empty", "Not_empty",
"Empty", "Empty", "Not_empty", "Not_empty", "Not_empty",
"Not_empty", "Empty", "Empty", "Empty", "Not_empty", "Not_empty",
"Not_empty", "Not_empty", "Empty", "Not_empty", "Not_empty",
"Empty", "Empty", "Empty", "Not_empty", "Not_empty", "Not_empty",
"Not_empty")), row.names = c(NA, -25L), class = c("data.table",
"data.frame"))
I looked online and various SO posts like 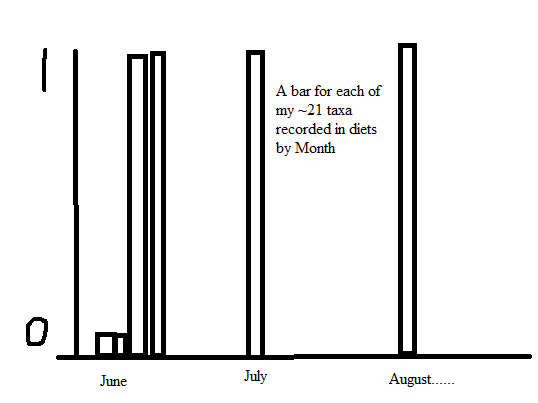
Originally, the data was in long format but this results in multiple rows per fish. I changed to wide format to end up with one observation(row) per fish.
How can I achieve this to ultimately plot presence/absence by month? Thank you!
CodePudding user response:
Maybe you want something like with your selected column converted to a longer format. After that to show the zeros bars, you can say that there is a bar by giving it a small negative number (if you want to show zero bars). At last, the y-axis has a binary format. You can use the following code:
library(dplyr)
library(ggplot2)
library(tidyr)
library(lubridate)
df %>%
select(MONTH, Daphnia, Byths, Chiro.Pupae, Isopod, Goby) %>%
mutate(MONTH = month.name[MONTH]) %>%
pivot_longer(cols = -c(MONTH), values_transform = as.numeric) %>%
ggplot(aes(x = MONTH, y = sapply(value, FUN=function(x) ifelse(x==0,-0.1,x)), fill = name))
geom_bar(position = "dodge", stat = "identity")
scale_y_continuous(breaks = c(0,1))
labs(y = "Absence", x = "Month")
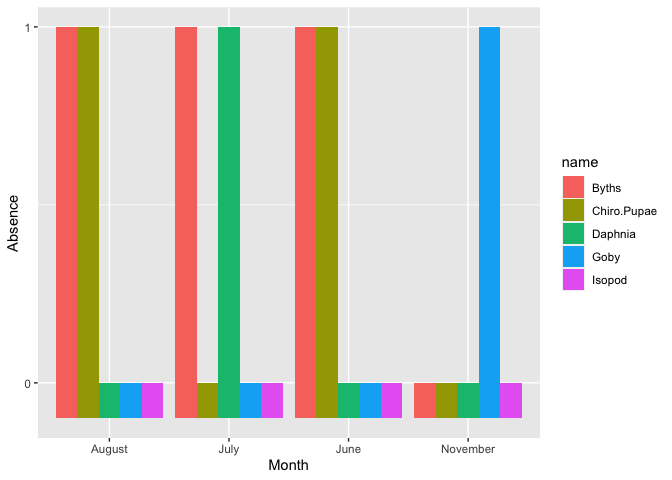
Created on 2022-07-30 by the reprex package (v2.0.1)
