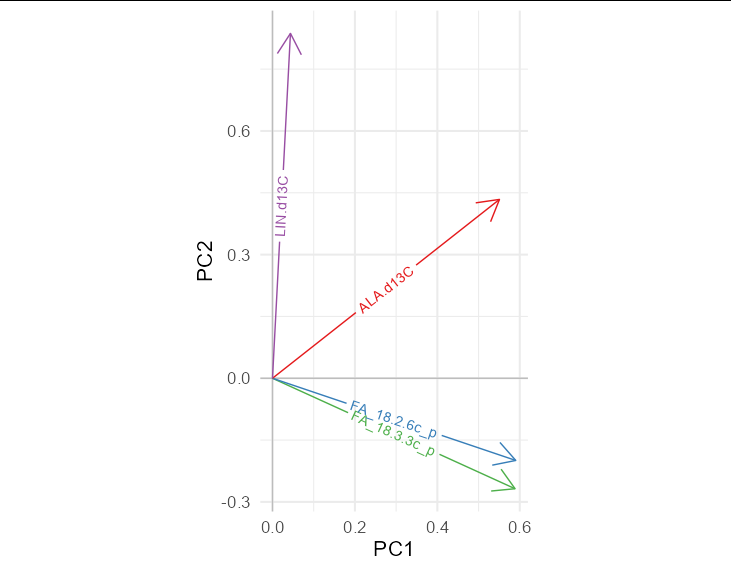
I want to plot arrows in a pca. I can do that with nmds. But for unknown reasons I cannot do it with the pca.
First I tried this:
library(AMR)
ggplot_pca(pca_resources)
The plot is nice but I want the groupings to be color-coded and I do not know how to do that here.
Then I tried it via ggplot2:
ggplot(PCi,aes(x=PC1,y=PC2,color=organ))
geom_point(size=2)
geom_abline(intercept=0,slope=0,lty=3,size=0.4)
geom_vline(xintercept=0,lty=3,size=0.4)
xlab("PC1 (53%)")
ylab("PC2 (33%)")
theme(#legend.justification=c(1,0),legend.position=c(0.9,0.5),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.text.y=element_text(size=15,family="Arial",color="black"),
axis.text.x=element_text(size=15,family="Arial",color="black"),
axis.title.x=element_text(size=15,family="Arial"),
axis.title.y=element_text(size=15,family="Arial"),
legend.title=element_text(size=20,family="Arial"),
legend.text=element_text(size=20,family="Arial"))
theme_bw()
Here are the PC scores for the arrows:
structure(c(0.588517849867411, 0.590285718836058, 0.55073730129091,
0.0435653014731059, -0.267996461049223, -0.199411444560057, 0.433923525535396,
0.836733736997915, 0.608676467420504, -0.768503848789056, 0.179696493593818,
-0.081387730002692, -0.459714688976792, -0.14560109997207, 0.690005774717737,
-0.539772873792367), .Dim = c(4L, 4L), .Dimnames = list(c("FA_18.3.3c_p",
"FA_18.2.6c_p", "ALA.d13C", "LIN.d13C"), c("PC1", "PC2", "PC3",
"PC4")))
Here are the data points for the PCA plot:
structure(list(PC1 = c(-1.88530472821989, -2.24714159937733,
-1.34432344539257, -0.968510753543999, -0.624879049572724, -2.36471016819961,
-1.16311229527896, -2.61987812001917, -2.08025468016444, -2.68276183422677
), PC2 = c(-1.50284890951331, -0.363086620597548, 1.45264977869589,
0.721195171897019, 0.93112605562114, -0.906913023282559, -0.283296924307472,
0.267889285983414, 1.33171541603416, -1.80603301970074), PC3 = c(-0.542337614825214,
0.646911614334602, 0.0237708210906419, 0.168460682596456, -0.82053283734023,
-0.101006562583037, -0.151718041641362, 0.574991815166341, 0.564555094889462,
-0.459509110124471), PC4 = c(0.0280582360577376, 0.863570266385183,
-0.466390733446598, 1.13230673371715, 0.269061697019256, -0.50980826233187,
0.181559477299738, -0.12716527942606, 0.107402240826891, -0.785580543274415
), organ = structure(c(4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L
), .Label = c("brain", "crustacea", "ephemeroptera", "epilithon",
"eyes", "fresh.leaves", "liver", "muscle", "plecoptera", "submerged.leaves"
), class = "factor")), row.names = c("3", "4", "5", "6", "7",
"8", "9", "10", "11", "13"), class = "data.frame")
How can I add the arrows its labels?
Thanks, Nadine
CodePudding user response:
You can reshape the PC co-ordinates and plot them as segments. Here I have used geom_textsegment from the geomtextpath package to easily label the arrows:
library(tidyverse)
library(geomtextpath)
PC_scores %>%
as.data.frame() %>%
select(1:2) %>%
rownames_to_column(var = "var") %>%
ggplot(aes(0, 0, color = var))
geom_hline(yintercept = 0, alpha = 0.2)
geom_vline(xintercept = 0, alpha = 0.2)
geom_textsegment(aes(xend = PC1, yend = PC2, label = var),
arrow = arrow())
scale_color_brewer(palette = "Set1")
theme_minimal(base_size = 16)
theme(legend.position = "none")
labs(x = "PC1", y = "PC2")
coord_equal()
CodePudding user response:
You could also use the package ggfortify like this:
PCi <- structure(list(PC1 = c(-1.88530472821989, -2.24714159937733,
-1.34432344539257, -0.968510753543999, -0.624879049572724, -2.36471016819961,
-1.16311229527896, -2.61987812001917, -2.08025468016444, -2.68276183422677
), PC2 = c(-1.50284890951331, -0.363086620597548, 1.45264977869589,
0.721195171897019, 0.93112605562114, -0.906913023282559, -0.283296924307472,
0.267889285983414, 1.33171541603416, -1.80603301970074), PC3 = c(-0.542337614825214,
0.646911614334602, 0.0237708210906419, 0.168460682596456, -0.82053283734023,
-0.101006562583037, -0.151718041641362, 0.574991815166341, 0.564555094889462,
-0.459509110124471), PC4 = c(0.0280582360577376, 0.863570266385183,
-0.466390733446598, 1.13230673371715, 0.269061697019256, -0.50980826233187,
0.181559477299738, -0.12716527942606, 0.107402240826891, -0.785580543274415
), organ = structure(c(4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L
), .Label = c("brain", "crustacea", "ephemeroptera", "epilithon",
"eyes", "fresh.leaves", "liver", "muscle", "plecoptera", "submerged.leaves"
), class = "factor")), row.names = c("3", "4", "5", "6", "7",
"8", "9", "10", "11", "13"), class = "data.frame")
library(ggfortify)
pca_res <- prcomp(PCi[,-5])
autoplot(pca_res, data = PCi, colour = 'organ', loadings = TRUE, loadings.label = TRUE)
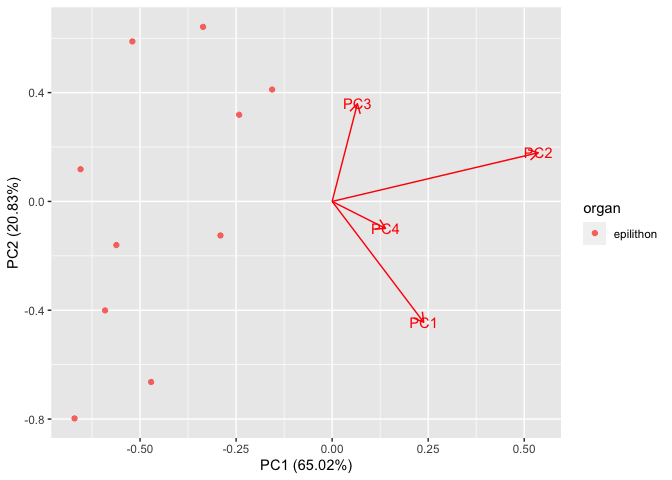
Created on 2022-07-30 by the reprex package (v2.0.1)