Any input/thoughts on this are appreciated.
import { cv } from 'https://deno.land/x/[email protected]/mod.ts';
export const match = (imagePath: string, templatePath: string) => {
const imageSource = Deno.readFileSync(imagePath);
const imageTemplate = Deno.readFileSync(templatePath);
const src = cv.matFromImageData({ data: imageSource, width: 640, height: 640 });
const templ = cv.matFromImageData({ data: imageTemplate, width: 29, height: 25 });
const processedImage = new cv.Mat();
const logResult = new cv.Mat();
const mask = new cv.Mat();
cv.matchTemplate(src, templ, processedImage, cv.TM_SQDIFF, mask);
cv.log(processedImage, logResult)
console.log(logResult.empty())
};
UPDATE
Using @ChristophRackwitz's answer and digging into opencv(js) docs, I managed to get close to my goal.
I decided to step down from taking multiple matches into account, and focused on a single (best) match of my needle in the haystack. Since ultimately this is my use-case anyways.
Going through the code provided in this example and comparing data with the data in my code, I figured something was off with the binary image data which I supplied to cv.matFromImageData. I solved this my properly decoding the png and passing that decoded image's bitmap to cv.matFromImageData.
I used TM_SQDIFF as suggested, and got some great results.
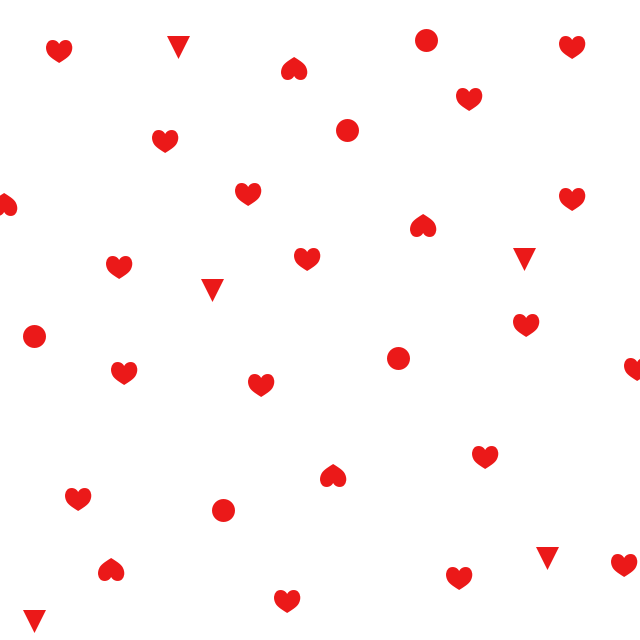
Haystack

Needle

Result

I achieved this in the following way.
import { cv } from 'https://deno.land/x/[email protected]/mod.ts';
import { Image } from 'https://deno.land/x/[email protected]/mod.ts';
export type Match = false | {
x: number;
y: number;
width: number;
height: number;
center?: {
x: number;
y: number;
};
};
export const match = async (haystackPath: string, needlePath: string, drawOutput = false): Promise<Match> => {
const perfStart = performance.now()
const haystack = await Image.decode(Deno.readFileSync(haystackPath));
const needle = await Image.decode(Deno.readFileSync(needlePath));
const haystackMat = cv.matFromImageData({
data: haystack.bitmap,
width: haystack.width,
height: haystack.height,
});
const needleMat = cv.matFromImageData({
data: needle.bitmap,
width: needle.width,
height: needle.height,
});
const dest = new cv.Mat();
const mask = new cv.Mat();
cv.matchTemplate(haystackMat, needleMat, dest, cv.TM_SQDIFF, mask);
const result = cv.minMaxLoc(dest, mask);
const match: Match = {
x: result.minLoc.x,
y: result.minLoc.y,
width: needleMat.cols,
height: needleMat.rows,
};
match.center = {
x: match.x (match.width * 0.5),
y: match.y (match.height * 0.5),
};
if (drawOutput) {
haystack.drawBox(
match.x,
match.y,
match.width,
match.height,
Image.rgbaToColor(255, 0, 0, 255),
);
Deno.writeFileSync(`${haystackPath.replace('.png', '-result.png')}`, await haystack.encode(0));
}
const perfEnd = performance.now()
console.log(`Match took ${perfEnd - perfStart}ms`)
return match.x > 0 || match.y > 0 ? match : false;
};
ISSUE
The remaining issue is that I also get a false match when it should not match anything.
Based on what I know so far, I should be able to solve this using a threshold like so:
cv.threshold(dest, dest, 0.9, 1, cv.THRESH_BINARY);
Adding this line after matchTemplate however makes it indeed so that I no longer get false matches when I don't expect them, but I also no longer get a match when I DO expect them.
Obviously I am missing something on how to work with the cv threshold. Any advice on that?
UPDATE 2
After experimenting and reading some more I managed to get it to work with normalised values like so:
cv.matchTemplate(haystackMat, needleMat, dest, cv.TM_SQDIFF_NORMED, mask);
cv.threshold(dest, dest, 0.01, 1, cv.THRESH_BINARY);
Other than it being normalised it seems to do the trick consistently for me. However, I would still like to know why I cant get it to work without using normalised values. So any input is still appreciated. Will mark this post as solved in a few days to give people the chance to discus the topic some more while it's still relevant.
CodePudding user response:
The TM_* methods are treacherous. And the docs throw formulas at you that would make anyone feel dumb, because they're code, not explanation.
Consider the calculation of one correlation: one particular position of the template/"needle" on the "haystack".
All the CCORR modes will simply multiply elementwise. Your data uses white as "background", which is a "DC offset". The signal, the difference to white of anything not-white, will drown in the large "DC offset" of your data. The calculated correlation coefficients will vary mostly with the DC offset and hardly at all with the actual signal/difference.
This is what that looks like, roughly. The result of running with TM_CCOEFF_NORMED, overlaid on top of the haystack (with some padding). You're getting big fat responses for all instances of all shapes, no matter their specific shape.
You want to use differences instead. The SQDIFF modes will handle that. Squared differences are a measure of dissimilarity, i.e. a perfect match will give you 0.
Let's look at some values...
(hh, hw) = haystack.shape[:2]
(nh, nw) = needle.shape[:2]
scores = cv.matchTemplate(image=haystack, templ=needle, method=cv.TM_SQDIFF)
(sh, sw) = scores.shape # will be shaped like haystack - needle
scores = np.log10(1 scores) # any log will do
maxscore = 255**2 * (nh * nw * 3)
# maximum conceivable SQDIFF score, 3-channel data, any needle
# for a specific needle:
#maxscore = (np.maximum(needle, 255-needle, dtype=np.float32)**2).sum()
# map range linearly, from [0 .. ~8] to [1 .. 0] (white to black)
(smin, smax) = (0.0, np.log10(1 maxscore))
(omin, omax) = (1.0, 0.0)
print("mapping from", (smin, smax), "to", (omin, omax))
out = (scores - smin) / (smax - smin) * (omax - omin) omin
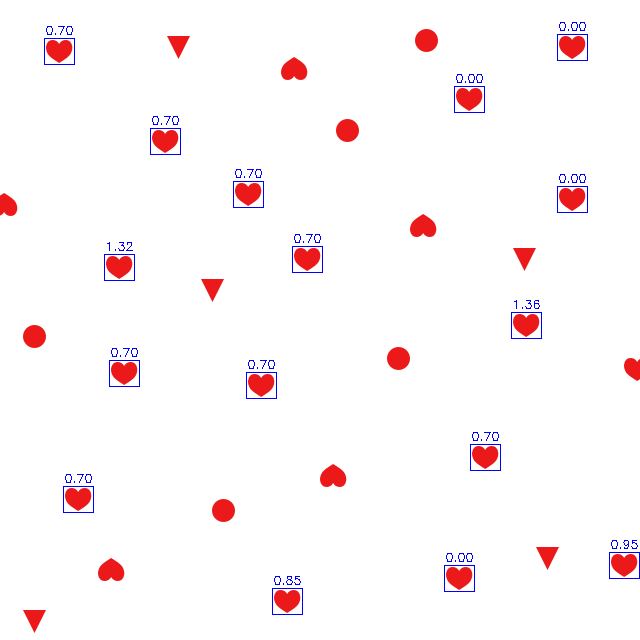
You'll see gray peaks, but some are actually (close to) white while others aren't. Those are truly instances of the needle image. The other instances differ more from the needle, so they're just some reddish shapes that kinda look like the needle.
Now you can find local extrema. There are many ways to do that. You'll want to do two things: filter by absolute value (threshold) and suppress non-maxima (scores above threshold that are dominated by better nearby score). I'll just do the filtering and pretend there aren't any nearby non-maxima because the resulting peaks fall off strongly enough. If that happens to not be the case, you'd see double drawing in the picture below, boxes becoming "bold" because they're drawn twice onto adjacent pixel positions.
I'm picking a threshold of 2.0 because that represents a difference of 100, i.e. one color value in one pixel may have differed by 10 (10*10 = 100), or two values may have differed by 7 (7*7 = 49, twice makes 98), ... so it's still a very tiny, imperceptible difference. A threshold of 6 would mean a sum of squared differences of upto a million, allowing for a lot more difference.
(i,j) = (scores <= 2.0).nonzero() # threshold "empirically decided"
instances = np.transpose([j,i]) # list of (x,y) points
That's giving me 16 instances.
canvas = haystack.copy()
for pt in instances:
(j,i) = pt
score = scores[i,j]
cv.rectangle(canvas,
pt1=(pt-(1,1)).astype(int), pt2=(pt (nw,nh)).astype(int),
color=(255,0,0), thickness=1)
cv.putText(canvas,
text=f"{score:.2f}",
org=(pt [0,-5]).astype(int),
fontFace=cv.FONT_HERSHEY_SIMPLEX, fontScale=0.4,
color=(255,0,0), thickness=1)
That's drawing a box around each, with the logarithm of the score above it.
One simple approach to get candidates for Non-Maxima Suppression (NMS) is to cv.dilate the scores and equality-compare, to gain a mask of candidates. Those scores that are local maxima, will compare equal to themselves (the dilated array), and every surrounding score will be less. This alone will have some corner cases you will need to handle. Note: at this stage, those are local maxima of any value. You need to combine (logical and) that with a mask from thresholding the values.
NMS commonly is required to handle immediate neighbors being above the threshold, and merge them or pick the better one. You can do that by simply running connectedComponents(WithStats) and taking the blob centers. I think that's clearly better than trying to find contours.
The dilate-and-compare approach will not suppress neighbors if they have the same score. If you did the connectedComponents step, you only have non-immediate neighbors to deal with here. What to do is up to you. It's not clear cut anyway.