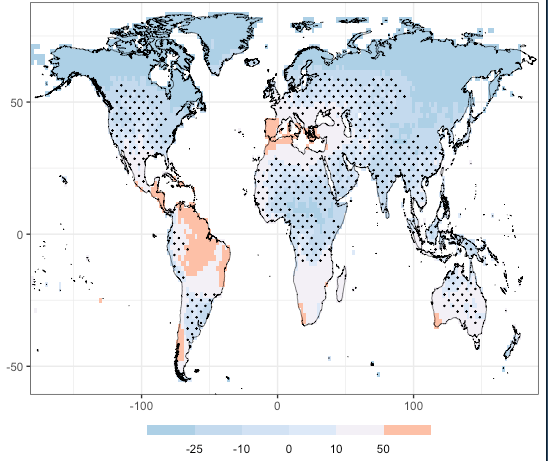
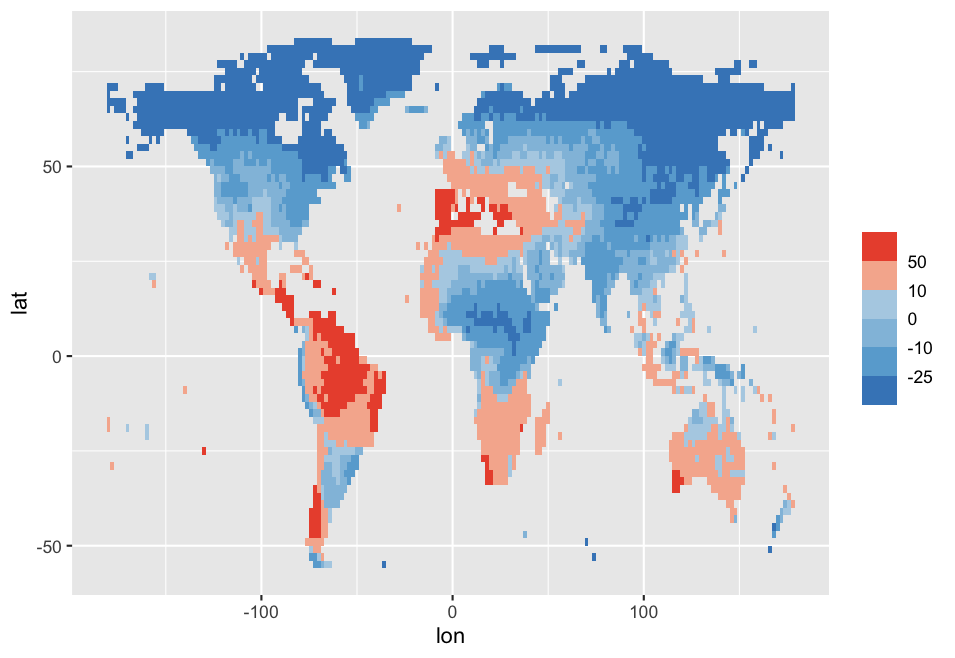
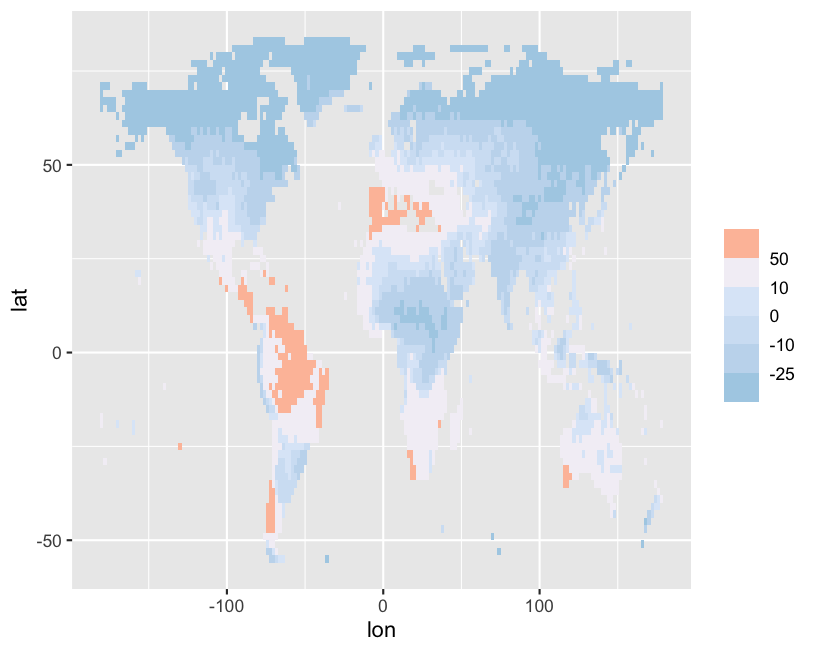
I am using scale_fill_stepsn() because I want a tailored colorbar in my geom_tile() plot.
Therefore, I choose 6 colors and 5 breaks as follows:
scale_fill_stepsn(name = "", colors=c("#9ecae1", # dark blue
"#c6dbef", # medium blue
"#eff3ff", # light blue
"#fee5d9", # dark red
"#fcbba1", # medium red
"#fc9272"), # light red
breaks = c(-25, -10, 0, 10, 50))
The color palette has 3 tones of blue and 3 of red, the values plotted with geom_tile() are both negative and positive, and hence the breaks().
Therefore, I am expecting that scale_fill_stepsn() will put the colors as follows:
#9ecae1 -25 #c6dbef -10 #eff3ff 0 #fee5d9 10 #fcbba1 50 #fc9272
But instead I get the following:
Any idea what is happing here?
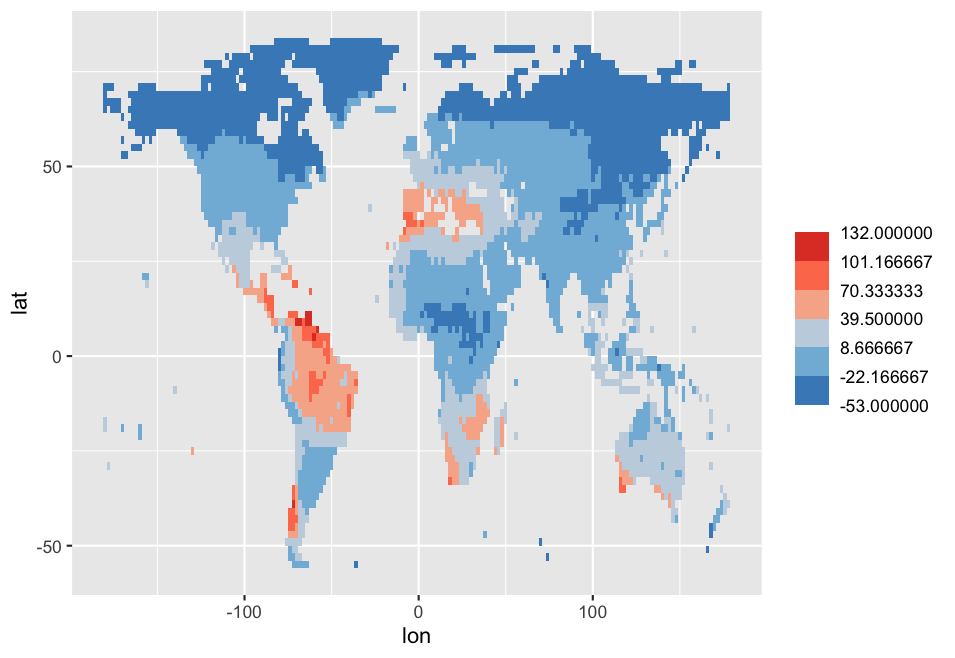
################# UPDATE
Edit2
Your breaks need to be set at your minimum and maximum values. You can create a seq of values with min and max. The values are used to map these values between 0 and 1. You need 1 more breaks because of the length of your palette, and that's why you get a grey color every time. You can use the following code:
plot_1 = ggplot()
geom_tile(data=test, aes(x=lon, y=lat, fill = median))
scale_fill_stepsn(name = "",
colors =c("#2171b5", "#6baed6", "#bdd7e7", "#fcae91", "#fb6a4a", "#cb181d"),
breaks = seq(min(test$median), max(test$median), length = 7),
values = scales::rescale(seq(min(test$median), max(test$median), length = 6)))
plot_1
Output:
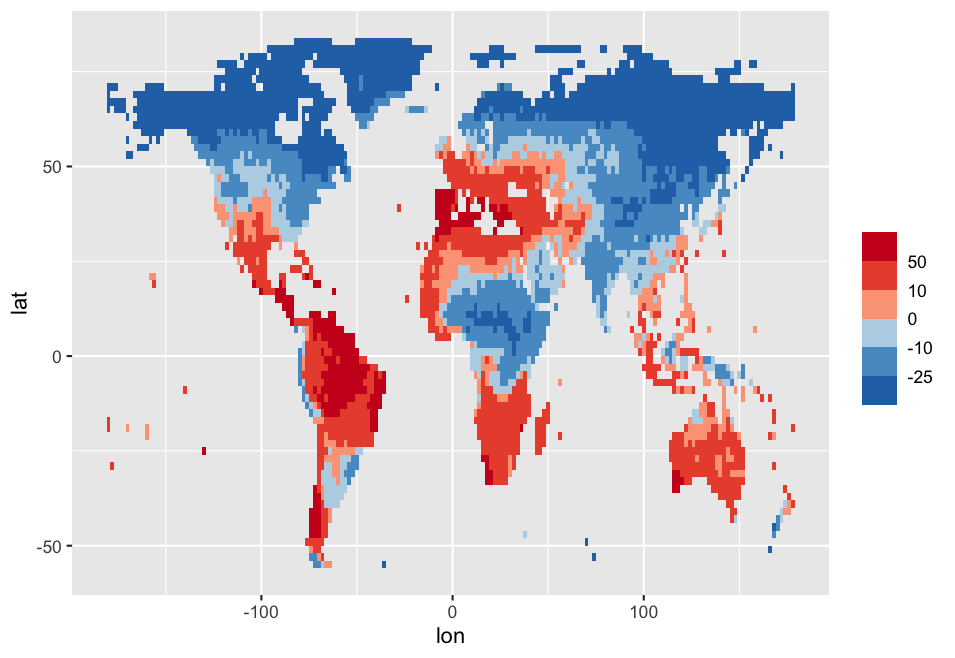
Edit1
You should mention the breaks in your values argument too with a rescale range between 0 and 1, because your colors are not evenly positioned. Here you can see what the documenation says about the argument values:
if colours should not be evenly positioned along the gradient this vector gives the position (between 0 and 1) for each colour in the colours vector. See rescale() for a convenience function to map an arbitrary range to between 0 and 1.
You could use rescale from scales package for that. Here is a reproducible example:
library(ggplot2)
plot_1 = ggplot()
geom_tile(data=test, aes(x=lon, y=lat, fill = median))
scale_fill_stepsn(name = "",
colors =c("#9ecae1", "#c6dbef", "#eff3ff", "#fee5d9", "#fcbba1", "#fc9272"),,
breaks = c(-25, -10, 0, 10, 50),
values = scales::rescale(c(-25, -10, 0, 10, 50)))
plot_1
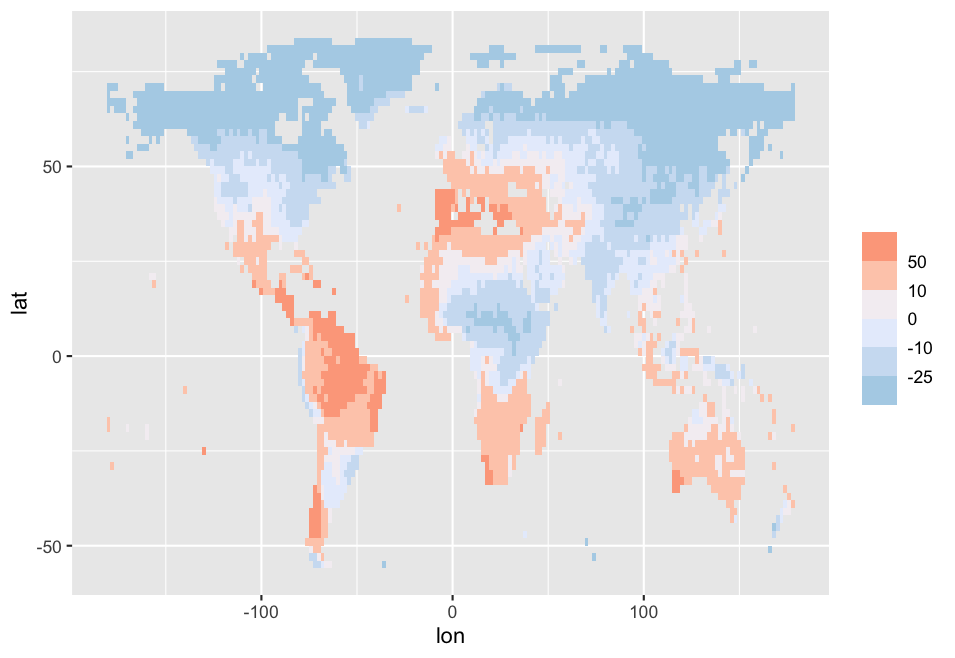
Output:
As you can see, now the colors are shown right!
Different colors:
library(ggplot2)
plot_1 = ggplot()
geom_tile(data=test, aes(x=lon, y=lat, fill = median))
scale_fill_stepsn(name = "",
colors =c("#2171b5", "#6baed6", "#bdd7e7", "#fcae91", "#fb6a4a", "#cb181d"),
breaks = c(-25, -10, 0, 10, 50),
values = scales::rescale(c(-25, -10, 0, 10, 25, 50)))
plot_1
Output:
Old answer
It seems that the hex code colors you use, are different than you think. You could use the package plotrix to check the color names of the hex codes. You can see it suggests something different below here:
library(ggplot2)
plot_1 = ggplot()
geom_tile(data=test, aes(x=lon, y=lat, fill = median))
scale_fill_stepsn(name = "",
colors = c("lightskyblue2", "lightsteelblue2", "aliceblue", "antiquewhite", "burlywood2", "salmon1"),
breaks = c(-25, -10, 0, 10, 50))
plot_1
library(plotrix)
sapply(c("#9ecae1", "#c6dbef", "#eff3ff", "#fee5d9", "#fcbba1", "#fc9272"), color.id)
#> #9ecae1 #c6dbef #eff3ff #fee5d9
#> "lightskyblue2" "lightsteelblue2" "aliceblue" "antiquewhite"
#> #fcbba1 #fc9272
#> "burlywood2" "salmon1"
Created on 2022-10-27 with reprex v2.0.2