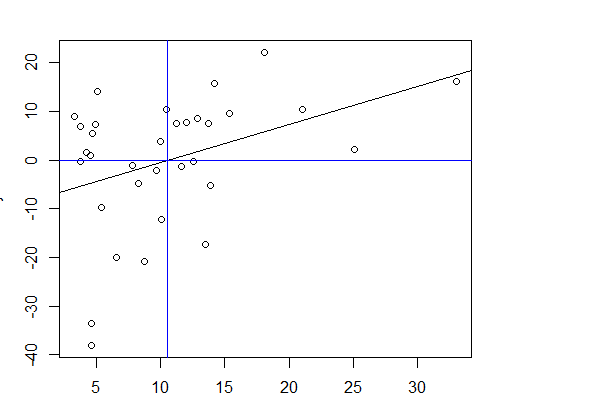
I am trying to calculate the value of x where y = 0. I could able to do it for single x using the following code
lm.model <- lm(y ~ x)
cc <- coef(lm.model)
f <- function(x) cc[2]*x cc[1]
plot(x, y)
abline(coef(lm.model))
abline(h=0, col="blue")
(threshold <- uniroot(f, interval = c(0, 100))$root)
abline(v=threshold, col="blue")
x = c(33.05, 14.22, 15.35, 13.52, 8.7, 13.73, 8.28, 21.02, 9.97,
11.98, 12.87, 5.05, 11.23, 11.65, 10.05, 12.58, 13.88, 9.66,
4.62, 4.56, 5.35, 3.7, 3.29, 4.87, 3.75, 6.55, 4.51, 7.77, 4.7,
4.18, 25.14, 18.08, 10.41)
y = c(16.22699279, 15.78620732, 9.656361014, -17.32805679, -20.85685895,
7.601993251, -4.776053714, 10.50972236, 3.853479771, 7.713563136,
8.579366561, 14.16989395, 7.484692081, -1.2807472, -12.13759458,
-0.29138513, -5.238157067, -2.033194068, -38.12157566, -33.61912493,
-9.763657548, -0.240863712, 9.090638907, 7.345492608, 6.949676888,
-19.94866471, 0.995659732, -1.162616185, 5.497998429, 1.656653092,
2.116687436, 22.23175649, 10.33039543)
But I have multiple x variables. Now how can I apply it for multiple x variables at a time? Here is an example data
df = structure(list(y = c(16.2269927925813, 15.7862073196372, 9.65636101412767,
-17.3280567922775, -20.8568589521297, 7.6019932507973, -4.77605371404423,
10.5097223644541, 3.85347977129367, 7.71356313645697, 8.57936656085966,
14.1698939499927, 7.4846920807874, -1.28074719969249, -12.1375945758837,
-0.291385130176774, -5.23815706681139, -2.03319406769161, -38.1215756639013,
-33.6191249261727, -9.76365754821171, -0.240863712421707, 9.09063890677045,
7.34549260800693, 6.94967688778232, -19.9486647079697, 0.995659731521127,
-1.16261618452931, 5.49799842947493, 1.65665309209479, 2.11668743610013,
22.2317564898722, 10.3303954315884), x1 = c(8.56, 8.66, 9.09,
8.36, 8.3, 8.63, 8.78, 8.44, 8.34, 8.46, 8.33, 8.19, 8.58, 8.65,
8.75, 8.34, 8.77, 9.06, 9.31, 9.11, 9.26, 9.81, 9.68, 9.79, 9.26,
9.53, 8.89, 8.89, 10.37, 9.58, 10.27, 10.16, 10.27), x2 = c(164,
328.3, 0, 590.2, 406.6, 188.4, 423.8, 355.3, 337.6, 0, 0, 200.1,
0, 315.8, 547.5, 225.6, 655.7, 387.2, 0, 487.4, 400.4, 0, 234.9,
275.5, 0, 0, 613.2, 207.4, 184.4, 162.8, 220, 174.8, 0), x3 = c(4517.7,
2953.4, 2899.3, 2573.8, 3310.7, 3880.3, 3016.8, 3552.3, 2960.1,
323, 2638.5, 3343.1, 3274.7, 3218, 3268.3, 3507.9, 3709.2, 3537.5,
2634.4, 1964.6, 3333.7, 2809.7, 3326.8, 3524.5, 3893.9, 3166.7,
3992.1, 4324.7, 3077.9, 3069.9, 4218.9, 3897.4, 2693.9), x4 = c(14.7,
14.5, 15.5, 17, 16.2, 15.9, 15.7, 15.3, 13.5, 14, 15.4, 16.2,
15.6, 15.7, 15.1, 15.8, 15.3, 14.9, 15.7, 16.3, 15.21000004,
16.7, 15.6, 16.2, 15.7, 16.3, 17.3, 16.9, 15.7, 14.9, 13.81999969,
14.90754509, 12.42847157), x5 = c(28.3, 29.1, 28.3, 29.1, 28.7,
29.3, 28.9, 28.4, 29.3, 29.3, 29.1, 29, 29.9, 29.5, 28.4, 30.3,
29.1, 29.1, 29, 29.5, 29.3, 28.5, 29, 28.7, 29.4, 28.8, 29.2,
30.1, 28.3, 28.7, 24.96999931, 25.79496384, 25.3072052), x6 = c(33.05,
14.22, 15.35, 13.52, 8.7, 13.73, 8.28, 21.02, 9.97, 11.98, 12.87,
5.05, 11.23, 11.65, 10.05, 12.58, 13.88, 9.66, 4.62, 4.56, 5.35,
3.7, 3.29, 4.87, 3.75, 6.55, 4.51, 7.77, 4.7, 4.18, 25.14, 18.08,
10.41), x7 = c(13.8425, 11.1175, 8.95, 13.5375, 5.4025, 13.5625,
13.735, 14.14, 8.0875, 5.565, 12.255, 3.3075, 6.345, 4.8125,
4.0325, 11.475, 10.32, 17.71, 2.3375, 3.92, 5.7, 2.42, 8.3075,
7.4725, 7.7925, 10.8725, 8.005, 11.7475, 13.405, 8.425, 47.155,
26.1, 6.6675), x8 = c(0.95, 3.01, 1.92, 1.51, 2.61, 1.32, 3.55,
1.21, 2.14, 1.1, 1.32, 0.76, 1.34, 5.41, 9.38, 6.55, 4.44, 7.37,
9.84, 12.68, 15.52, 23.01, 18.59, 21.64, 19.69, 25.22, 22.38,
25.03, 37.42, 22.26, 2.1, 3.01, 0.82), x9 = c(26.2, 25.8, 25.8,
25.5, 26, 24.7, 22.9, 25.3, 26.3, 26.1, 22.5, 25.9, 26.4, 25.2,
25.8, 25.4, 25, 23.2, 26.4, 25.8, 26.6, 26.2, 25.8, 26.8, 25,
25.4, 25.6, 26.1, 25.7, 25.8, 24.78000069, 24.98148918, 26.39899826
), x10 = c(35.4, 39, 37.5, 36.4, 37.1, 36.2, 37.3, 36.4, 37.5,
36, 36.6, 35.6, 37.3, 38.3, 37, 37.5, 37.5, 39.6, 37.8, 36.8,
36.6, 38.4, 38.9, 38.4, 38.4, 37.7, 39.1, 37.7, 37.8, 39.4, 36.25,
35.57029343, 35.57416534), x11 = c(653.86191565, 383.1, 457.1,
591.4, 549.2, 475.2, 626.4, 308.8, 652.4, 77, 380.9, 530.5, 393,
712.1, 623.4, 515.7, 706.4, 713.4, 343.7, 559.5, 630.1, 292.3,
578.6, 628.88904574, 480.96959685, 591.35600287, 804.8, 419.6,
403.7, 361.2, 515.07101438, 434.66682808, 299.9531298), x12 = c(163.9793854,
167.9, 135, 215.8, 213, 188.4, 260.6, 191.8, 337.6, 55, 147.6,
200.1, 140.7, 315.8, 189.6, 225.6, 469.3, 201.8, 140, 297.2,
204.6, 142.5, 234.9, 275.494751, 153.7796173, 147.6174622, 433.6,
207.4, 184.4, 162.8, 219.9721832, 174.8355713, 106.8163605),
x13 = c(92, 67, 67, 50, 70, 87, 68, 86, 70, 11, 66, 79, 70,
61, 75, 78, 78, 77, 69, 35, 72, 76, 69, 84, 93, 73, 81, 99,
80, 76, 101, 86, 80), x14 = c(70, 42, 46, 34, 55, 60, 51,
65, 49, 1, 40, 56, 54, 41, 48, 57, 46, 50, 41, 22, 47, 47,
49, 57, 70, 52, 56, 70, 48, 50, 74, 66, 47), x15 = c(21,
12, 13, 10, 14, 16, 10, 13, 10, 0, 9, 14, 16, 20, 14, 14,
13, 15, 10, 7, 17, 8, 14, 14, 14, 11, 17, 19, 12, 11, 17,
17, 9), x16 = c(1076.8, 783.7, 711.8, 1041.9, 957.4, 939.3,
662.9, 768.1, 770.3, 0, 399.2, 606.2, 724.1, 960.8, 943.8,
737.8, 1477.4, 1191.7, 371.3, 956.4, 1251.7, 345.7, 1210.7,
845, 598.1, 821.7, 1310.6, 940.1, 581, 520, 313.5, 606.8,
201.2), x17 = c(163.9793854, 167.9, 128.4, 215.8, 213, 188.4,
260.6, 191.8, 337.6, 55, 147.6, 200.1, 140.7, 315.8, 189.6,
225.6, 469.3, 201.8, 140, 297.2, 204.6, 142.5, 234.9, 157.7472534,
153.7796173, 147.6174622, 133.1873627, 150.2, 184.4, 162.8,
219.9721832, 174.8355713, 106.8163605)), row.names = c(NA,
33L), class = "data.frame")
CodePudding user response:
You can use purrr::map to loop through every x.
library(dplyr)
library(purrr)
thresholds <- df %>%
select(-y) %>%
map_dbl(function(x){
lm.model <- lm(df$y ~ x)
cc <- coef(lm.model)
f <- function(x) cc[2]*x cc[1]
plot(x, df$y)
abline(coef(lm.model))
abline(h=0, col="blue")
threshold <- tryCatch(uniroot(f, interval = c(0, 100))$root, error = function(cond){NA})
abline(v=threshold, col="blue")
return(threshold)})
For some x's, uniroot(f, interval = c(0, 100))$root yields an error: Error
in uniroot(f, interval = c(0, 100)) : f() values at end points not of opposite sign
So the tryCatch is used to return NA for the threshold associated with that x, instead of breaking the code.
Result:
> thresholds
x1 x2 x3 x4 x5 x6 x7 x8 x9
9.023314 NA NA 15.459841 28.727293 10.514728 10.493577 9.669244 25.522480
x10 x11 x12 x13 x14 x15 x16 x17
37.370852 NA NA 73.398380 50.239522 13.022176 NA NA
Edit: binding the graphs together
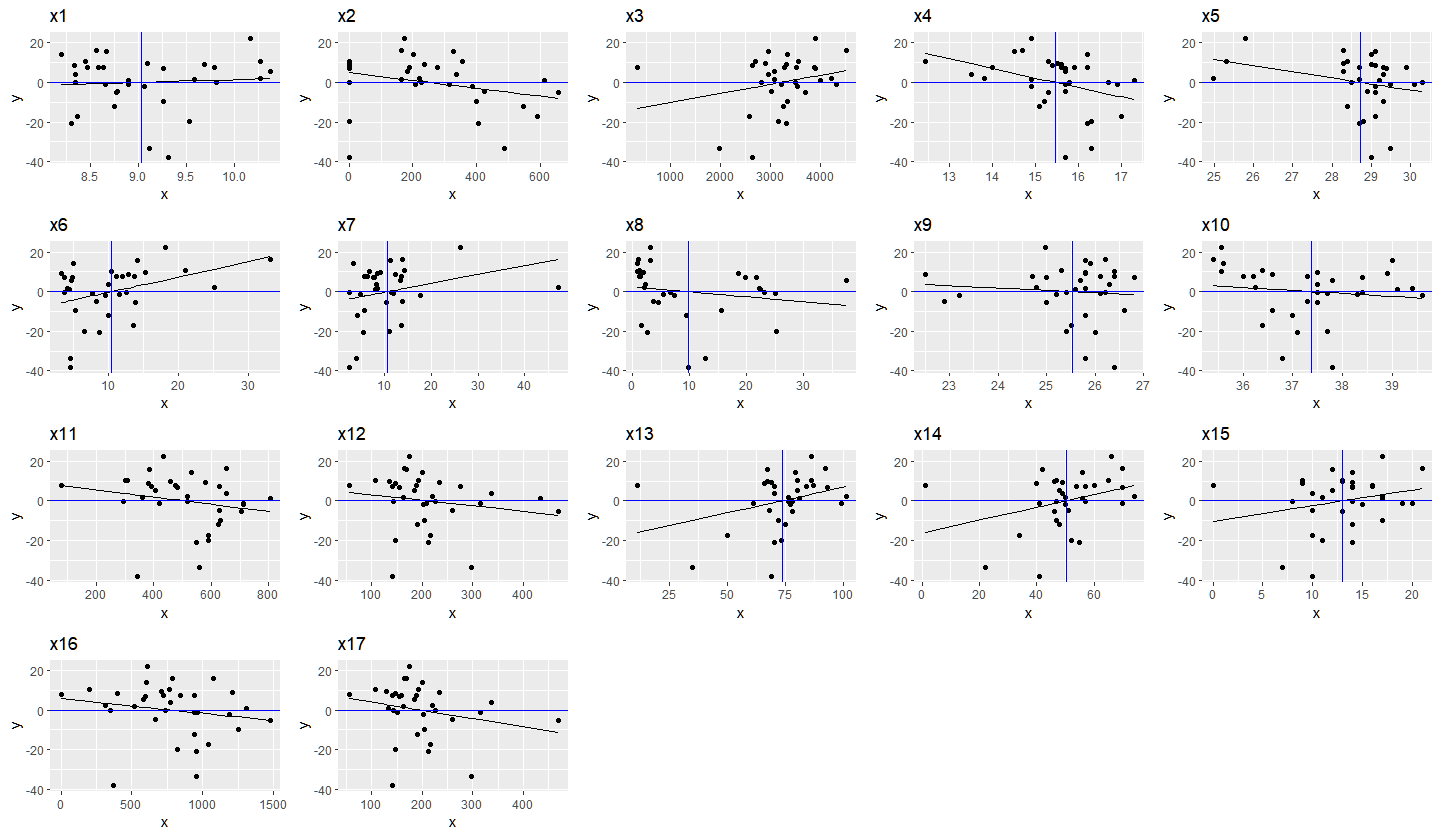
graphs <- df %>%
select(-y) %>%
imap(function(x, name){
lm.model <- lm(df$y ~ x)
cc <- coef(lm.model)
f <- function(x) cc[2]*x cc[1]
threshold <- tryCatch(uniroot(f, interval = c(0, 100))$root, error = function(cond){NA})
g = ggplot(mapping = aes(x))
geom_point(aes(y = df$y))
geom_line(aes(y = cc[2]*x cc[1]))
geom_hline(yintercept = 0, color = "blue")
labs(title = name, y = "y", x = "x")
if(!is.na(threshold)) {g = g geom_vline(xintercept = threshold, color = "blue")}
return(g)})
ggpubr::ggarrange(plotlist = graphs)
Result:
Obs2: i assumed that you don't need the thresholds vector defined in the first attempt, if you still need it, it's easy to add it back to the answer
Obs1: let me know if you want any aesthetic change on the graphs
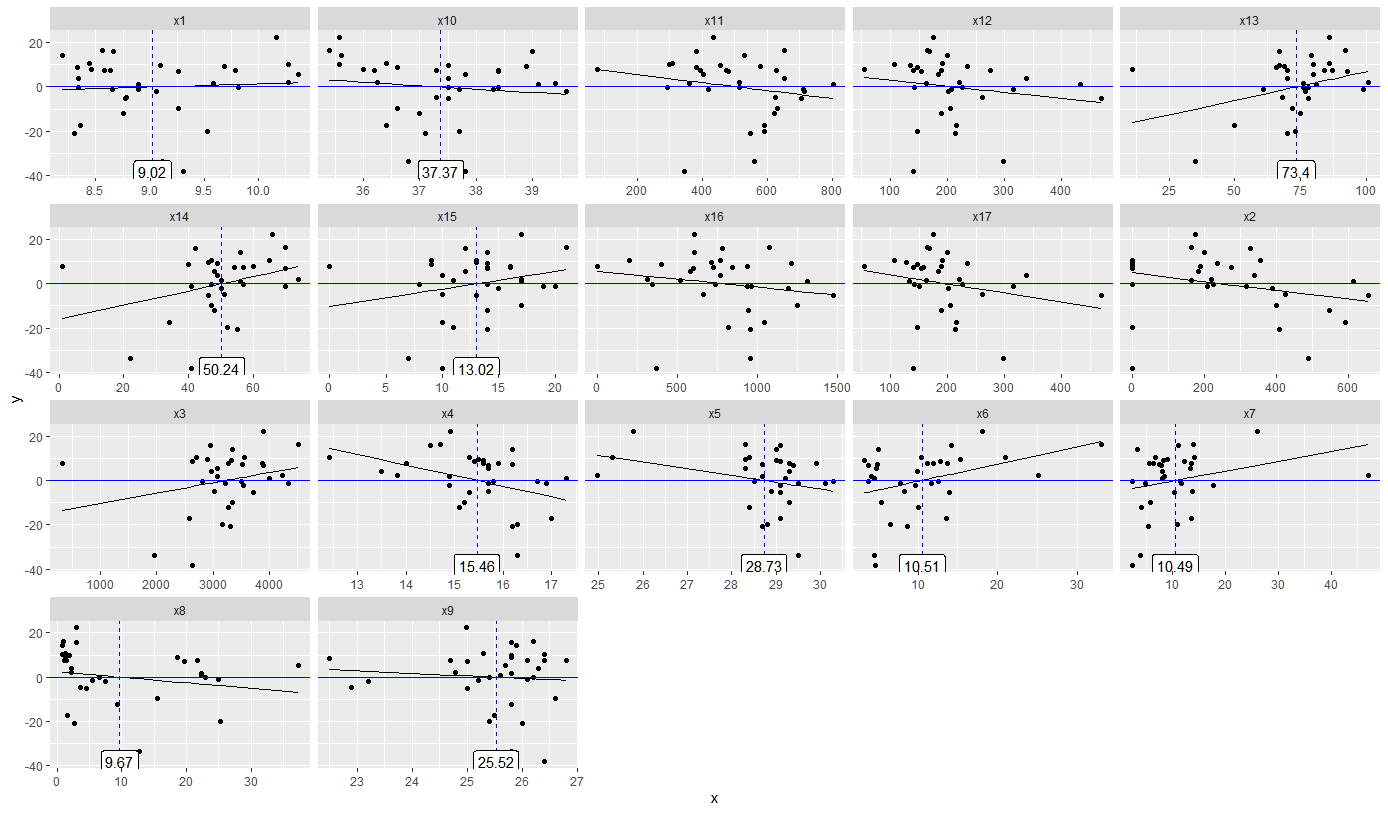
Edit 2: graph with common axis
To use a common axis is better to use facets instead of ggarrange. In order to do that, we need to first save the fitted data for all variables, then plot, so the ggplot expression goes out of the map. Also, we now save the treshold info.
graphs <- df %>%
select(-y) %>%
imap(function(x, name){
lm.model <- lm(df$y ~ x)
cc <- coef(lm.model)
f <- function(x) cc[2]*x cc[1]
threshold <- tryCatch(uniroot(f, interval = c(0, 100))$root, error = function(cond){NA})
list(threshold = threshold,
data = tibble(y = df$y, "name" = name, "x" = x, "fitted" = cc[2]*x cc[1]))})
Now we use the purrr::transpose() function to build a dataset for the data and other for the treshold. This functions does something like:
list(x1 = list(treshold, data), x2 = ...) >>> list(treshold = list(x1, x2, ...), data = list(x1, x2, ...))
df2 = graphs %>%
transpose() %>%
`$`(data) %>%
bind_rows() %>%
mutate(name = factor(name, paste0("x", 1:17)))
thresholds = graphs %>%
transpose() %>%
`$`(threshold) %>%
{tibble(int = as.numeric(.), name = names(.))} #both datasets have the name column, to be used inside `facet_wrap()`
ggplot(df2, aes(x))
geom_point(aes(y = y))
geom_line(aes(y = fitted))
facet_wrap(vars(name), scales = "free_x")
geom_hline(yintercept = 0, color = "blue")
geom_vline(aes(xintercept = int), thresholds, color = "blue", linetype = 2)
geom_label(aes(label = round(int, 2), x = int*1, y = min(df$y)), thresholds, size = 4)
Result:
Obs1: the labels position and size can be easily altered. Another option is using the thresholds as a axis break
Obs2: this method can be slow for large datasets. A more efficient option is to save only threshold and cc inside map, and then building the dataset after it.