How do I get the new hclust object after using cutree function? I know the standard output from cutree is a numerical vector that tells you the clusters each leaf node belongs. However, how can we get the aggregated tree structure after cutting at a certain height or by a given number of clusters?
library(gplots)
library(dendextend)
data <- iris[,1:4]
distance <- dist(data, method = "euclidean", diag = FALSE, upper = FALSE)
hc <- hclust(distance, method = 'ward.D')
dnd <- as.dendrogram(hc)
plot(dnd)
abline(h=hc$height[140],col="red")
I searched related topics, but unfortunately cant find a solution.
CodePudding user response:
Try this:
library(dplyr)
numberGroup = 10
dataClust = cbind(data, clust = cutree(hc, numberGroup))
dataClust = dataClust %>%
group_by(clust) %>%
summarise_all(mean)
dataClust %>%
dist %>%
hclust("ward.D2") %>%
plot
CodePudding user response:
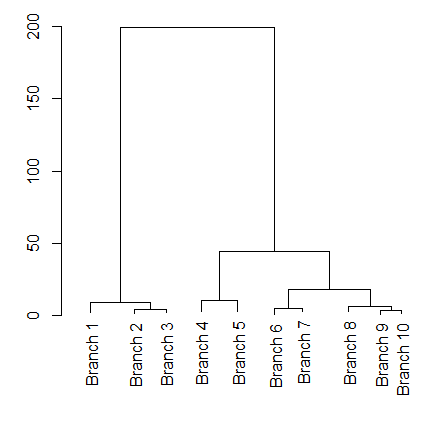
You can use cut to get the pieces of the dendrogram. Since you seem to be interested in the height h=hc$height[140]
CutDend = cut(dnd, h=hc$height[140])
plot(CutDend$upper)
BTW, you can also get the 10 branches below the cut line using CutDend$lower
Some additional details are available via help(cut.dendrogram)
cut.dendrogram() returns a list with components $upper and $lower, the first is a truncated version of the original tree, also of class dendrogram, the latter a list with the branches obtained from cutting the tree, each a dendrogram.