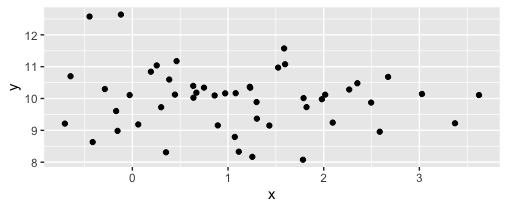
Having a tibble and a simple scatterplot:
p <- tibble(
x = rnorm(50, 1),
y = rnorm(50, 10)
)
ggplot(p, aes(x, y)) geom_point()
I get something like this:
I would like to align (center, left, right, as the case may be) the title of the x-axis - here rather blandly x - with a specific value on the axis, say the off-center 0 in this case. Is there a way to do that declaratively, without having to resort to the dumb (as in "free of context") trial-and-error element_text(hjust=??). The ?? are rather appropriate here because every value is a result of experimentation (my screen and PDF export in RStudio never agree on quite some plot elements). Any change in the data or the dimensions of the rendering may (or may not) invalidate the hjust value and I am looking for a solution that graciously repositions itself, much like the axes do.
Following the suggestions in the comments by @tjebo I dug a little deeper into the coordinate spaces. hjust = 0.0 and hjust = 1.0 clearly align the label with the Cartesian coordinate system extent (but magically left-aligned and right-aligned, respectively) so when I set specific limits, calculation of the exact value of hjust is straightforward (aiming for 0 and hjust = (0 - -1.5) / (3.5 - -1.5) = 0.3):
ggplot(p, aes(x, y))
geom_point()
coord_cartesian(ylim = c(8, 12.5), xlim = c(-1.5, 3.5), expand=FALSE)
theme(axis.title.x = element_text(hjust = 0.3))
This gives an acceptable result for a label like x, but for longer labels the alignment is off again:
ggplot(p %>% mutate(`Longer X label` = x), aes(x = `Longer X label`, y = y))
geom_point()
coord_cartesian(ylim = c(8, 12.5), xlim = c(-1.5, 3.5), expand=FALSE)
theme(axis.title.x = element_text(hjust = 0.3))
Any further suggestions much appreciated.
CodePudding user response:
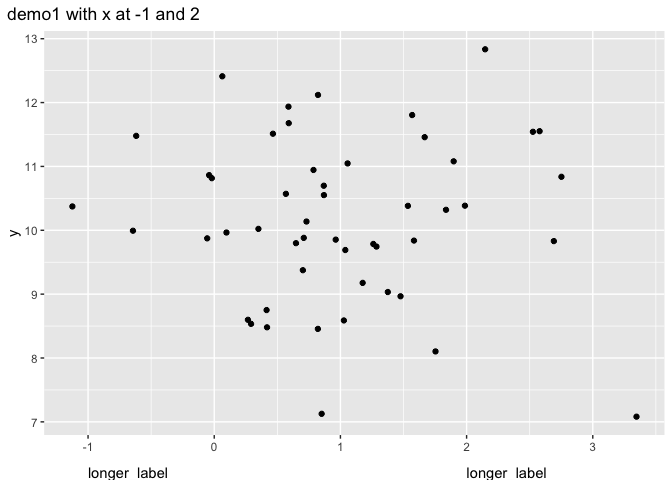
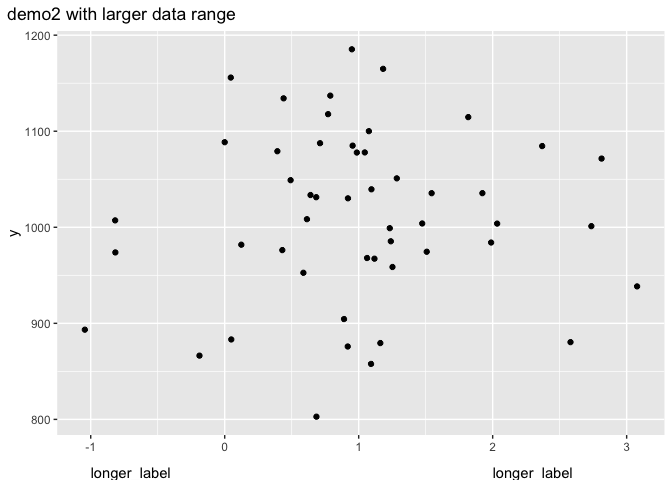
Another option (different enough hopefully to justify the second answer) is as already mentioned to create the annotation as a separate plot. This removes the range problem. I like {patchwork} for this.
library(tidyverse)
library(patchwork)
p <- tibble( x = rnorm(50, 1), y = rnorm(50, 10))
p1 <- tibble( x = rnorm(50, 1), y = 100*rnorm(50, 10))
## I like to define constants outside my ggplot call
mylab <- "longer_label"
x_demo <- c(-1, 2)
demo_fct <- function(p){
p1 <- ggplot(p, aes(x, y))
geom_point()
labs(x = NULL)
theme(plot.margin = margin())
p2 <- ggplot(p, aes(x, y))
## you need that for your correct alignment with the first plot
geom_blank()
annotate(geom = "text", x = x_demo, y = 1,
label = mylab, hjust = 0)
theme_void()
# you need that for those annoying margin reasons
coord_cartesian(clip = "off")
p1 / p2 plot_layout(heights = c(1, .05))
}
demo_fct(p) plot_annotation(title = "demo1 with x at -1 and 2")
demo_fct(p1) plot_annotation(title = "demo2 with larger data range")
Created on 2021-12-04 by the reprex package (v2.0.1)
CodePudding user response:
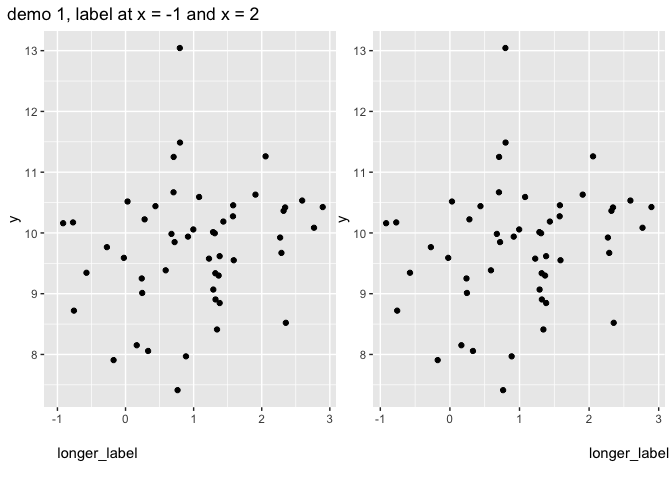
I still think you will fair better and easier with custom annotation. There are typically two ways to do that. Either direct labelling with a text layer (for single labels I prefer annotate(geom = "text"), or you create a separate plot and stitch both together, e.g. with patchwork.
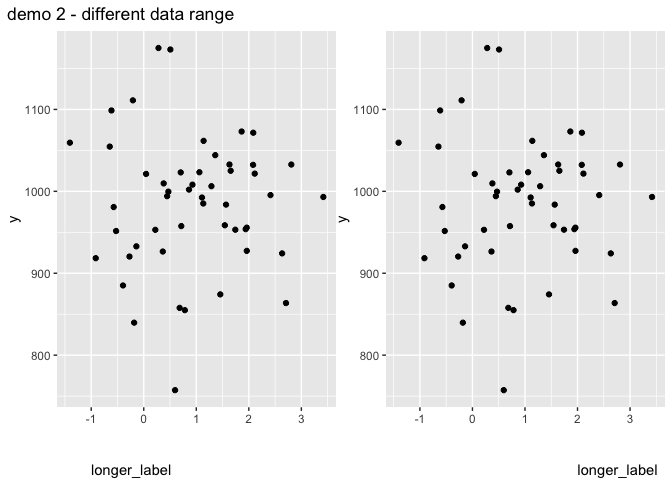
The biggest challenge is the positioning in y dimension. For this I typically take a semi-automatic approach where I only need to define one constant, and set the coordinates relative to the data range, so changes in range should in theory not matter much. (they still do a bit, because the panel dimensions also change). Below showing examples of exact label positioning for two different data ranges (using the same constant for both)
library(tidyverse)
# I only need patchwork for demo purpose, it is not required for the answer
library(patchwork)
p <- tibble( x = rnorm(50, 1), y = rnorm(50, 10))
p1 <- tibble( x = rnorm(50, 1), y = 100*rnorm(50, 10))
## I like to define constants outside my ggplot call
y_fac <- .1
mylab <- "longer_label"
x_demo <- c(-1, 2)
demo_fct <- function(df, x) {map(x_demo,~{
## I like to define constants outside my ggplot call
ylims <- range(df$y)
ggplot(df, aes(x, y))
geom_point()
## set hjust = 0 for full positioning control
annotate(geom = "text", x = ., y = min(ylims) - y_fac*mean(ylims),
label = mylab, hjust = 0)
coord_cartesian(ylim = ylims, clip = "off")
theme(plot.margin = margin(b = .5, unit = "in"))
labs(x = NULL)
})
}
demo_fct(p, x_demo) %>% wrap_plots() plot_annotation(title = "demo 1, label at x = -1 and x = 2")
demo_fct(p1, x_demo) %>% wrap_plots() plot_annotation(title = "demo 2 - different data range")
Created on 2021-12-04 by the reprex package (v2.0.1)