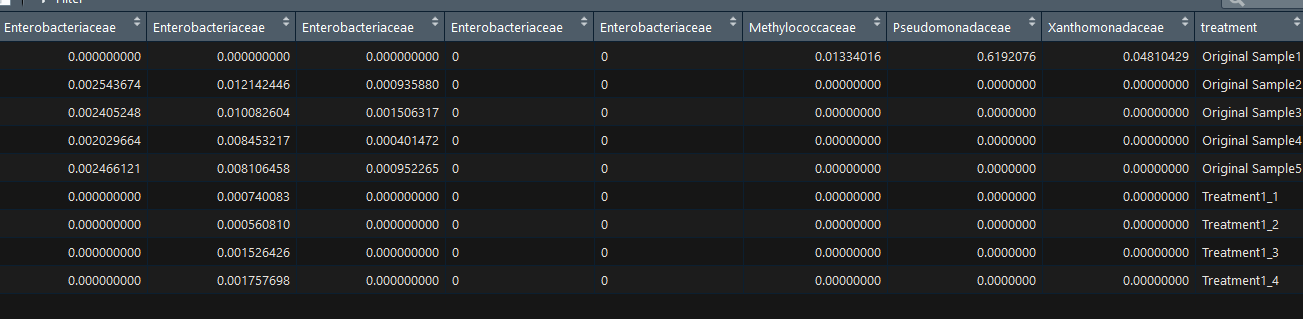
I have data that looks like this:
I want to plot columns with the same name, for example: Enterobacteriaceae treatment 1 together.
So it will look like this: x line - will contain the treatments: treatment 1_1 treatment 1_2 and so on. The y line will contain the values. Also, I would like to add the median and linear regression line.
The problem is that I keep getting an error since there are multiple columns with the same name and R sees that as a problem for plotting multiple columns with same name together.
What should I do? Should I try to merge columns with the same name?
CodePudding user response:
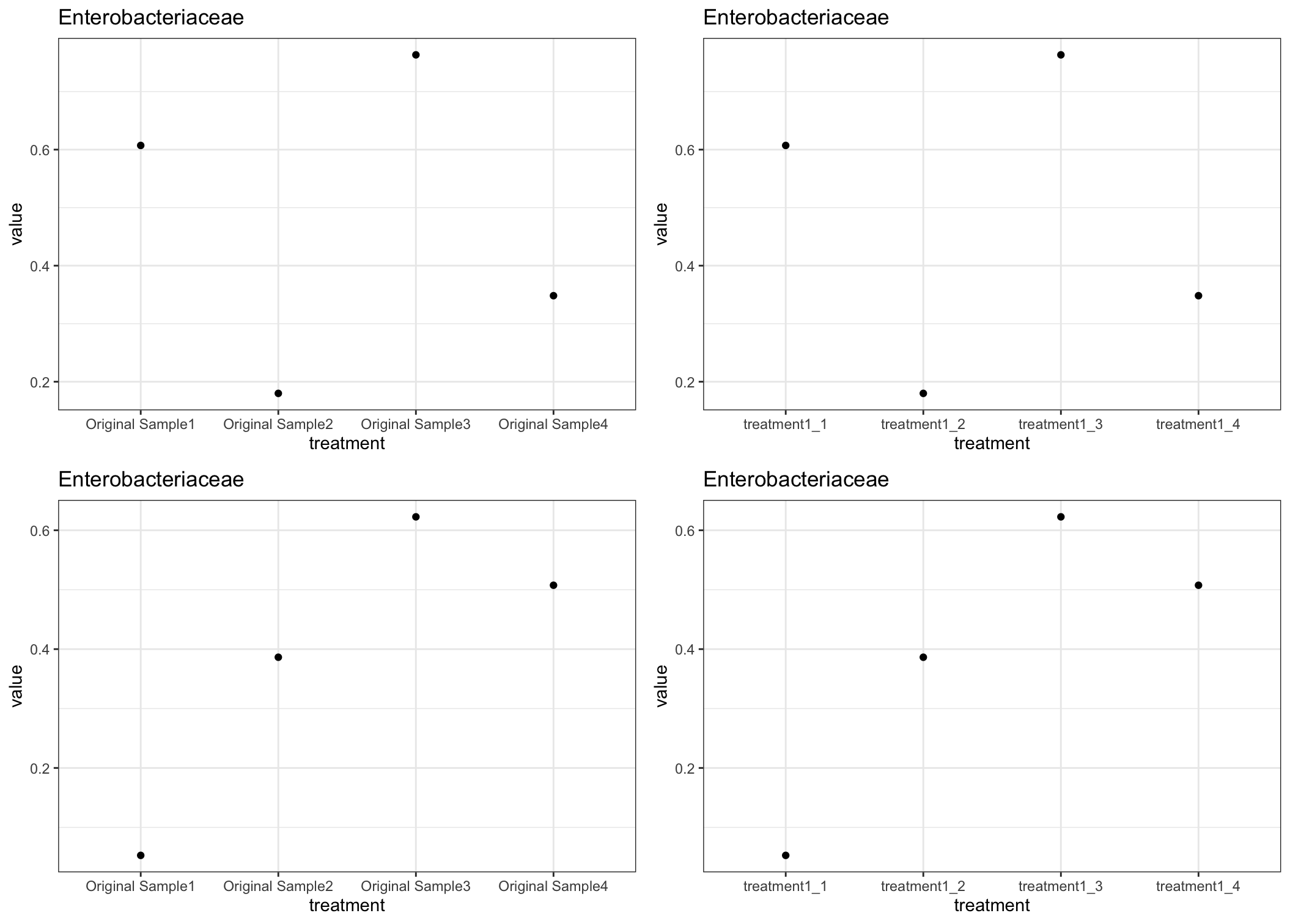
For plotting each group and column individually, you can put each into a nested list, so that we can take advantage of purrr functions. Then, create ggplot objects for each dataframe.
library(tidyverse)
library(ggpubr)
# First, split all columns into separate dataframes.
c_df <- df %>%
map(function(x)
as.data.frame(x)) %>%
# Then, you can bind the treatment column back to those dataframes.
map(function(x)
cbind(x, df$treatment)) %>%
# Remove "treatment" dataframe.
head(-1) %>%
# Then, split the original from treatment dataframes.
purrr::map(function(x)
split(x, f = str_detect(df$treatment, "treatment1")))
# Getting the names of the taxon (i.e., original column heading).
taxa_names <- names(c_df) %>%
rep(each = 2)
# Flatten list.
c_df <- c_df %>%
purrr::flatten() %>%
# Rename the 2 column names in all dataframes.
map( ~ .x %>%
dplyr::rename(value = "x", treatment = "df$treatment"))
# Replace the list names with the taxon names.
names(c_df) <- taxa_names
# Create a plotting function.
plot_treatment <- function(z, n) {
ggplot(data = z, aes(x = treatment, y = value))
geom_point()
theme_bw()
ggtitle(n)
}
# Use the plotting function to create all of the ggplot objects.
all_plots <- c_df %>%
purrr::map2(.y = names(c_df), .f = plot_treatment)
# Can plot in one figure.
ggarrange(all_plots[[1]],
all_plots[[2]],
all_plots[[3]],
all_plots[[4]],
ncol = 2,
nrow = 2)
Output (example)
Data
df <-
structure(
list(
Enterobacteriaceae = c(
0.60720596,
0.17991846,
0.76333618,
0.34825876,
0.60720596,
0.17991846,
0.76333618,
0.34825876
),
Enterobacteriaceae = c(
0.05291531,
0.38634377,
0.622598,
0.50749286,
0.05291531,
0.38634377,
0.622598,
0.50749286
),
Enterobacteriaceae = c(
0.3861723,
0.466643,
0.83439861,
0.99024876,
0.3861723,
0.466643,
0.83439861,
0.99024876
),
Methylococcaceae = c(
0.49516461,
0.16735156,
0.77037345,
0.50080786,
0.49516461,
0.16735156,
0.77037345,
0.50080786
),
Methylococcaceae = c(
0.18810595,
0.7514854,
0.05479668,
0.11263293,
0.18810595,
0.7514854,
0.05479668,
0.11263293
),
treatment = c(
"Original Sample1",
"Original Sample2",
"Original Sample3",
"Original Sample4",
"treatment1_1",
"treatment1_2",
"treatment1_3",
"treatment1_4"
)
),
class = "data.frame",
row.names = c(NA,-8L)
)
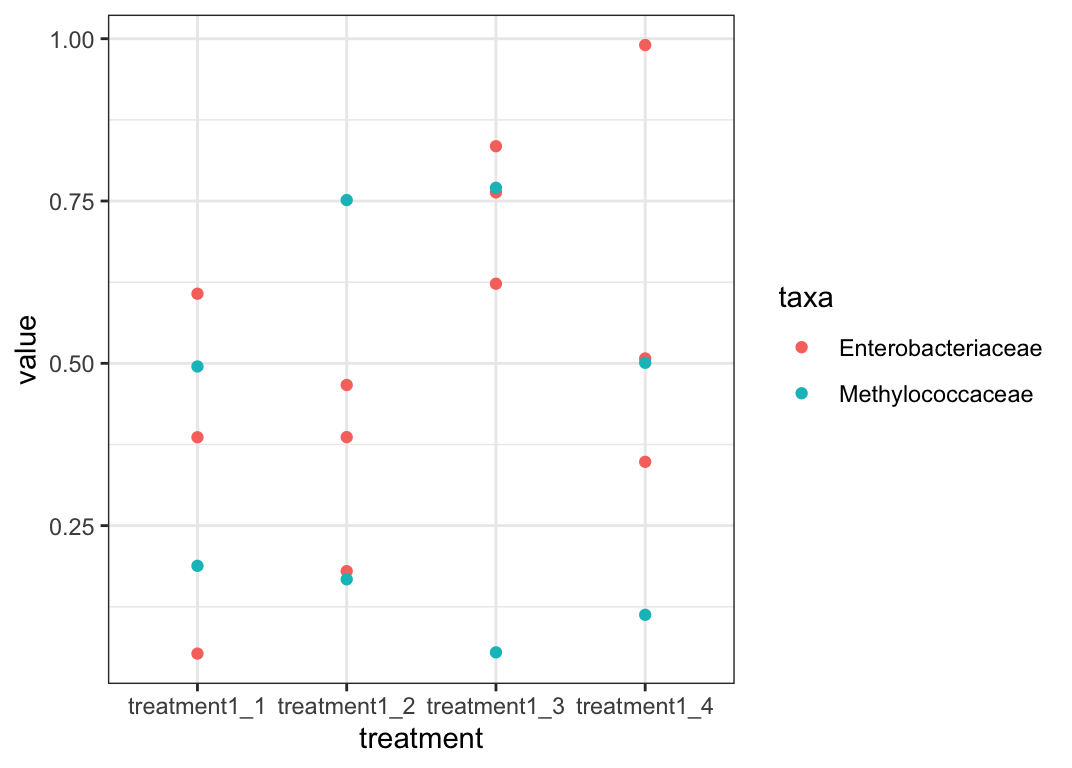
Generally, with ggplot, it is easiest to convert data into long format, which allows you to plot by groups. I created some dummy data as example. I am still unsure if this is the output you are looking for though.
library(tidyverse)
df %>%
tidyr::pivot_longer(!treatment, names_to = "taxa", values_to = "value") %>%
# You can change this to "Original" to get the other plot.
dplyr::filter(str_detect(treatment, "treatment1")) %>%
ggplot(aes(x = treatment, y = value, color = taxa))
geom_point()
theme_bw()
Output
Data
df <-
structure(
list(
Enterobacteriaceae = c(0.60720596, 0.17991846, 0.76333618, 0.34825876),
Enterobacteriaceae = c(0.05291531, 0.38634377, 0.622598, 0.50749286),
Enterobacteriaceae = c(0.3861723, 0.466643, 0.83439861, 0.99024876),
Methylococcaceae = c(0.49516461, 0.16735156, 0.77037345, 0.50080786),
Methylococcaceae = c(0.18810595, 0.7514854, 0.05479668, 0.11263293),
treatment = c(
"treatment1_1",
"treatment1_2",
"treatment1_3",
"treatment1_4"
)
),
class = "data.frame",
row.names = c(NA,-4L)
)