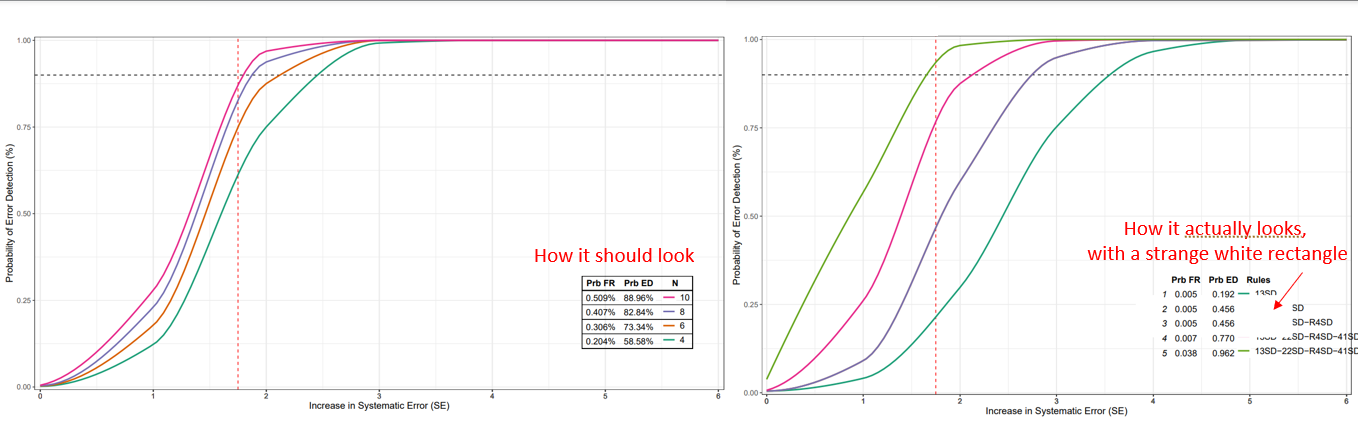
For the life of me, I cannot figure out why that rectangle is there or how to remove it. Any insight would be much appreciated.
Code:
library(dplyr)
library(ggplot2)
library(gridExtra)
library(grid)
library(patchwork)
#Data
fullData <- structure(list(error = c(0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4, 5, 6, 0, 1, 2, 3, 4,
5, 6), prob.ed = c(0.005, 0.041, 0.298, 0.753, 0.966, 0.998,
0.999, 0.005, 0.091, 0.598, 0.949, 0.997, 0.998, 0.999, 0.005,
0.091, 0.598, 0.949, 0.997, 0.998, 0.999, 0.007, 0.261, 0.875,
0.996, 1, 1, 1, 0.038, 0.568, 0.983, 1, 1, 1, 1), prob.fr = c(0.005,
0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005,
0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005, 0.005,
0.005, 0.005, 0.007, 0.007, 0.007, 0.007, 0.007, 0.007, 0.007,
0.038, 0.038, 0.038, 0.038, 0.038, 0.038, 0.038), N = c(2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2), rules = c("13SD", "13SD",
"13SD", "13SD", "13SD", "13SD", "13SD", "13SD-22SD", "13SD-22SD",
"13SD-22SD", "13SD-22SD", "13SD-22SD", "13SD-22SD", "13SD-22SD",
"13SD-22SD-R4SD", "13SD-22SD-R4SD", "13SD-22SD-R4SD", "13SD-22SD-R4SD",
"13SD-22SD-R4SD", "13SD-22SD-R4SD", "13SD-22SD-R4SD", "13SD-22SD-R4SD-41SD",
"13SD-22SD-R4SD-41SD", "13SD-22SD-R4SD-41SD", "13SD-22SD-R4SD-41SD",
"13SD-22SD-R4SD-41SD", "13SD-22SD-R4SD-41SD", "13SD-22SD-R4SD-41SD",
"13SD-22SD-R4SD-41SD-Xbar", "13SD-22SD-R4SD-41SD-Xbar", "13SD-22SD-R4SD-41SD-Xbar",
"13SD-22SD-R4SD-41SD-Xbar", "13SD-22SD-R4SD-41SD-Xbar", "13SD-22SD-R4SD-41SD-Xbar",
"13SD-22SD-R4SD-41SD-Xbar")), row.names = c(NA, -35L), class = "data.frame")
#Table for tableGrob
critTable <- structure(list(error = c(7, 7, 7, 7, 7), prob.ed = c(0.192, 0.456,
0.456, 0.77, 0.962), prob.fr = c(0.005, 0.005, 0.005, 0.007,
0.038), N = c(2, 2, 2, 2, 2), rules = c("13SD", "13SD-22SD",
"13SD-22SD-R4SD", "13SD-22SD-R4SD-41SD", "13SD-22SD-R4SD-41SD-Xbar"
)), row.names = c(NA, -5L), class = "data.frame")
#ggplot
plot <- plot <- ggplot(fullData, aes(x = error, y = prob.ed, group = rules, colour = as.factor(rules)))
geom_vline(xintercept = 1.75, colour = "red", linetype = "dashed")
geom_hline(yintercept = 0.90, linetype = "dashed")
geom_line(data = full.data %>%
group_by(rules) %>%
do({tibble(error = seq(min(.$error), max(.$error),length.out=100),
prob.ed = pchip(.$error, .$prob.ed, error))}),
size = 1)
scale_x_continuous(labels = full.data$error, breaks = full.data$error, expand = c(0, 0.05))
scale_y_continuous(expand = expansion(add = c(0.01, 0.01)))
scale_color_brewer(palette = "Dark2")
theme_bw()
theme(
legend.position=c(0.93,0.2075),
legend.title=element_blank(),
legend.background = element_blank(),
legend.key.size = unit(0.687, "cm"),
legend.text = element_text(size = 12))
#tableGrob
table <- tableGrob(critTable %>%
select(prob.fr,
prob.ed,) %>%
rename(`Prb FR` = prob.fr,
`Prb ED` = prob.ed) %>%
mutate(Rules = ""),
rows = NULL,
theme = ttheme_minimal())
#ggplot tableGrob
plot <- plot inset_element(table,
left = 0.65, bottom = 0.2, right = 0.9, top = .3, align_to = 'full')
CodePudding user response:
I adapted the code a little (added: library(pracma))
changed full.data to fullData in some positions in ggplot. The solution is not exactly the desired output. But it should be a good start to interfere.
So the white rectangle is caused by ttheme_minimal in tableGrob. You can adapt with gridExtra::ttheme_default().