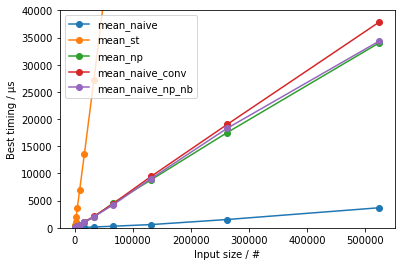
Was fooling around about what is the best way to calculate a mean of a list in python. Although I thought that numpy as optimized My results show that you shouldn't use numpy when it comes to this. I was wondering why and how python achieve this performance.
So basically I am trying to figure out how come native python is faster than numpy.
My code for testing:
import random
import numpy as np
import timeit
def average_native(l):
return sum(l)/len(l)
def average_np(l):
return np.mean(l)
def test_time(func, arg):
starttime = timeit.default_timer()
for _ in range(500):
func(arg)
return (timeit.default_timer() - starttime) / 500
for i in range(1, 7):
numbers = []
for _ in range(10**i):
numbers.append(random.randint(0, 100))
print("for " str(10**i) " numbers:")
print(test_time(average_native, numbers))
print(test_time(average_np, numbers))
The results:
for 10 numbers:
2.489999999999992e-07
8.465800000000023e-06
for 100 numbers:
8.554000000000061e-07
1.3220000000000009e-05
for 1000 numbers:
7.2817999999999495e-06
6.22666e-05
for 10000 numbers:
6.750499999999993e-05
0.0005553966000000001
for 100000 numbers:
0.0006954238
0.005352444999999999
for 1000000 numbers:
0.007034196399999999
0.0568878216
BTW I was running same code in c and was surprised to see that the python code is faster. test code:
#include <iostream>
#include <cstdlib>
#include <vector>
#include <chrono>
float calculate_average(std::vector<int> vec_of_num)
{
double sum=0;
uint64_t cnt=0;
for(auto & elem : vec_of_num)
{
cnt ;
sum = sum elem;
}
return sum / cnt;
}
int main()
{
// This program will create same sequence of
// random numbers on every program run
std::vector<int> vec;
for(int i = 0; i < 1000000; i )
vec.push_back(rand());
auto start = std::chrono::high_resolution_clock::now();
for(int i = 0; i < 500; i )
calculate_average(vec);
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double, std::milli> float_ms = end - start;
std::cout << "calculate_average() elapsed time is " << float_ms.count()/500 << " ms )" << std::endl;
return 0;
}
results:
calculate_average() elapsed time is 11.2082 ms )
Am I missing something?
Edit: I was running the c code on an online compiler (probebly without any optimization). Also it isn't the same Hardware, and how know what is going on it that server. After running and compiling the code in my device the code is much faster.
Edit2: So I changed the code for a numpy array in the numpy function and we do see that for smaller array/list the native python is better, however after around 1000 values numpy is preforming better. I don't really understand why. which optimizations numpy have that produce these results?
new results:
for 10 numbers:
2.4540000000000674e-07
6.722200000000012e-06
for 100 numbers:
8.497999999999562e-07
6.583400000000017e-06
for 1000 numbers:
6.990799999999964e-06
7.916000000000034e-06
for 10000 numbers:
6.61604e-05
1.5475799999999985e-05
for 100000 numbers:
0.0006671193999999999
8.412259999999994e-05
for 1000000 numbers:
0.0068192092
0.0008199298000000005
Maybe I need to restart this question :)
CodePudding user response:
C is much much slower than it needs to be.
First, for the C code, you're copying the vector, which is probably what's taking most of the time. You want to write:
float calculate_average(const std::vector<int>& vec_of_num)
instead of
float calculate_average(std::vector<int> vec_of_num)
In order to avoid making the copy.
Second, make sure you've compiled with optimizations on.
For the numpy version, you're doing an extra conversion, which slows you down.
and with:
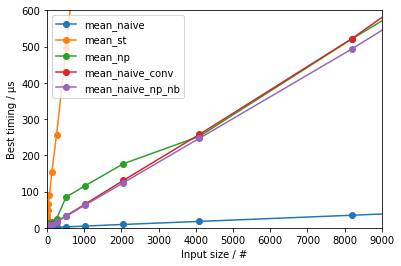
df.plot(marker='o', xlabel='Input size / #', ylabel='Best timing / µs', ylim=[0, 600], xlim=[0, 9000])
fig = plt.gcf()
fig.patch.set_facecolor('white')
(for some zooming on the smaller input sizes)
we can observe:
- The
statistics-based approach is the slowest by far and large - The naïve approach is the fastest by far and large
- When comparing the all the methods that do have a type conversion from Python
listto NumPy array:np.mean()is the fastest for larger input sizes, likely because it is compiled with specific optimizations (I'd speculate making optimal use of SIMD instructions); for smaller inputs, the running time is dominated by the overhead for supporting allufuncfunctionalities- the Numba-accelerated version is the fastest for medium input sizes; for very small inputs, the running time is lengthened by the small, roughly constant, overhead of calling a Numba function
- for very small inputs the
sum() / len()eventually gets to be the fastest
This indicates that sum() / len() is essentially slower than optimized C code acting on arrays.
CodePudding user response:
You are copying the array for each call to average which takes a lot of extra time.
#include <numeric>
#include <iostream>
#include <vector>
#include <chrono>
#include <random>
//!! pass vector by reference to avoid copies!!!!
double calculate_average(const std::vector<int>& vec_of_num)
{
return static_cast<double>(std::accumulate(vec_of_num.begin(), vec_of_num.end(), 0)) / static_cast<double>(vec_of_num.size());
}
int main()
{
std::mt19937 generator(1); // static std::mt19937 generator(std::random_device{}());
std::uniform_int_distribution<int> distribution{ 0,1000 };
// This program will create same sequence of
// random numbers on every program run
std::vector<int> values(1000000);
for (auto& value : values)
{
value = distribution(generator);
}
auto start = std::chrono::high_resolution_clock::now();
double sum{ 0.0 };
for (int i = 0; i < 500; i )
{
// force compiler to use average so it can't be optimized away
sum = calculate_average(values);
}
auto end = std::chrono::high_resolution_clock::now();
std::chrono::duration<double, std::milli> float_ms = end - start;
// force compiler to use sum so it can't be optimized away
std::cout << "sum = " << sum << "\n";
std::cout << "calculate_average() elapsed time is " << float_ms.count() / 500 << " ms )" << std::endl;
return 0;
}