I've searched around StackOverflow for the issue I'm facing and can't quite find something similar.
I'm working with a large time series, with a portion of the dataset below. With that, I'm trying to find a way to add an exponential fit to it using ggplot. Others have used geom_smooth(method = "lm", formula = (y ~ exp(x))) but that doesn't work with time series data or POSIXct class variables and returns the error "Computation failed in stat_smooth(): NA/NaN/Inf in 'x'". Previously, I simply used method = "loess", span = 0.1, but it doesn't capture the nature of the data very well.
Any help you could provide would be greatly appreciated!
data<-structure(list(avg_time = structure(c(1551420000, 1551506400,
1551592800, 1551679200, 1551765600, 1551852000, 1551938400, 1552024800,
1552111200, 1552197600, 1552280400, 1552366800, 1552453200, 1552539600,
1552626000, 1552712400, 1552798800, 1552885200, 1552971600, 1553058000,
1553144400, 1553230800, 1553317200, 1553403600, 1553490000, 1553576400,
1553662800, 1553749200, 1553835600, 1553922000, 1554008400, 1554094800,
1554181200, 1554267600, 1554354000, 1554440400, 1554526800, 1554613200,
1554699600, 1554786000, 1554872400, 1554958800, 1555045200, 1555131600,
1555218000, 1555304400, 1555390800, 1555477200, 1555563600, 1555650000,
1555736400, 1555822800, 1555909200, 1555995600, 1556082000, 1556168400,
1556254800, 1556341200, 1556427600, 1556514000, 1556600400, 1556686800,
1556773200, 1556859600, 1556946000, 1557032400, 1557118800, 1557205200,
1557291600, 1557378000, 1557464400, 1557550800, 1557637200, 1557723600,
1557810000, 1557896400, 1557982800, 1558069200, 1558155600, 1558242000,
1558328400, 1558414800, 1558501200, 1558587600, 1558674000, 1558760400,
1558846800, 1558933200, 1559019600, 1559106000, 1559192400, 1559278800,
1559365200, 1559451600, 1559538000, 1559624400, 1559710800), tzone = "", class = c("POSIXct",
"POSIXt")), ChlaMed = c(7.49786224129294, 6.33265484668835, 8.02891354394607,
8.36583527788548, 7.21848200004542, 3.87836804380364, 6.12041645730209,
6.11129053757413, 3.82314913061958, 6.66935722139803, 10.5846145945807,
1.3922819262622, 2.46397555374784, 3.5387541991258, 9.4377648342203,
3.8359888625491, 9.92938437268906, 9.84931346445947, 7.61136832417625,
10.422317215878, 9.92795625389519, 10.2145441518957, 9.87188069822321,
6.75768698400432, 7.50045495545547, 7.3979513362914, 12.0524471187313,
11.0031790178811, 9.23929610466274, 12.2253404703908, 10.8260865574934,
5.79312487695101, 7.86859910828088, 13.9784098169617, 13.3707820039944,
8.11038273190177, 13.852156279962, 6.94197529427832, 10.1752314872054,
10.3435349795235, 14.4105077850521, 12.3100928225917, 11.4965118440029,
13.5176883961026, 10.4577799463301, 11.8074169933709, 13.245655700942,
13.5716513275785, 14.0549071116729, 14.6034112846714, 13.8998981372714,
11.0290734663967, 12.7725741301044, 14.0037640681163, 12.99276716795,
12.9177278644427, 15.6103759408624, 11.4159351143177, 14.7053508114725,
14.3380030612979, 14.846661975045, 14.1918024501013, 14.1478311220769,
15.4169566103641, 14.1251696199414, 13.4057098254015, 15.0936022765442,
14.94796281727, 11.9943525040373, 15.6886181916423, 15.7057435474498,
16.1855936444667, 17.4195546581076, 16.977113306558, 16.4826655395595,
14.273959862613, 18.6570604979906, 15.2969835201503, 15.6502935625097,
16.4619111787213, 17.8995674961064, 16.9938925321631, 17.409705465615,
19.7838080835222, 18.7386731671602, 19.6515930205419, 20.4308399460097,
18.787235170191, 18.758368516805, 19.2927499812326, 19.4763785903839,
20.4249755976496, 19.0471858942877, 20.0134726662527, 20.9237871993584,
20.0967875761179, 20.7116516016657)), row.names = c(NA, -97L), class = c("tbl_df",
"tbl", "data.frame"))
CodePudding user response:
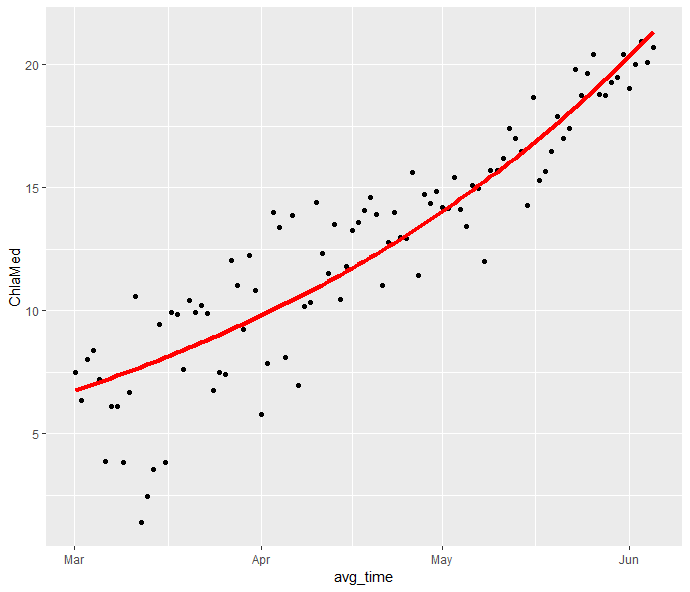
You could use nls() to get an exponential fit, make predictions, and plot those in addition to the raw data points:
data %>%
mutate(
d = as.numeric(difftime(as.Date(avg_time),min(as.Date(avg_time)),units = "days")),
preds =predict(nls(ChlaMed~a*exp(r*d), start = list(a=0.5, r=0.1), data=data))
) %>%
ggplot(aes(x=avg_time))
geom_point(aes(y=ChlaMed))
geom_line(aes(y=preds),color="red", linewidth=1.5)
CodePudding user response:
You can give it a try the timetk package using the natural log function for the response variable.
library(timetk)
data %>%
plot_time_series_regression(
.date_var = avg_time,
.formula = log(ChlaMed) ~ avg_time,
.interactive =FALSE
)