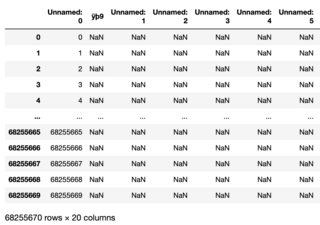
I have a .txt file that I am attempting to read in pandas. When I open the .txt file, I see it has the content and data I expect. However, when I read the file in pandas, the data is missing and I only NaNs.
here's sample content from .txt file:
980145115 189699454 SD Vacant Land Agricultural/Horticultural/Forest Vacant Land 3290522 216200 43.585481 -96.626588 10255 46099 I
707951172 189699522 AZ Government, Special Purpose Religious 91630 26730 102-55-008 4013 I
I have tried different parameters of encoding and sep in read_csv.
import pandas as pd
df = pd.read_csv('s3://filepath', encoding='latin-1', sep="\t")
Is there anything else I can try to read the data?
CodePudding user response:
It's probably due to the separator you have choosen in pandas.read_csv.
Try to use whitespaces instead with sep="\s\s " :
df = pd.read_csv('s3://filepath', encoding='latin-1', sep="\s\s ", engine="python", header=None)
Or with delim_whitespace=True :
df = pd.read_csv('s3://filepath', encoding='latin-1', delim_whitespace=True, header=None)
CodePudding user response:
The short answer
I would change two things to your call to read_csv:
- pass the
header=Noneargument, - pass the
na_filter=Falseargument.
df = pd.read_csv('s3://filepath', encoding='latin-1', sep="\t", header=None, na_filter=False)
Details
I copied your two sample data lines in a file called file.tsv on my computer.
No headers
You say your file does not contain a header, and by default read_csv() tries to interpret the first line as column headers. Just doing that, I get NaNs only for fields that were empty in your sample:
>>> pd.read_csv('file.tsv', encoding='latin-1', sep="\t", header=None)
0 1 2 3 4 5 6 7 8 ... 10 11 12 13 14 15 16 17 18
0 980145115 189699454 NaN NaN SD NaN NaN NaN Vacant Land ... NaN NaN 3290522 216200 43.585481 -96.626588 10255 46099 I
1 707951172 189699522 NaN NaN AZ NaN NaN NaN Government, Special Purpose ... NaN NaN 91630 26730 NaN NaN 102-55-008 4013 I
No NaN filter
The manual for pd.read_csv() says that na_filter=True is the default, and that means logic is applied to detect missing values. If that's not useful for you, and you just want to keep empty fields as empty values in your DF, turn that off:
>>> pd.read_csv('file.tsv', encoding='latin-1', sep="\t", header=None, na_filter=False)
0 1 2 3 4 5 6 7 8 ... 10 11 12 13 14 15 16 17 18
0 980145115 189699454 SD Vacant Land ... 3290522 216200 43.585481 -96.626588 10255 46099 I
1 707951172 189699522 AZ Government, Special Purpose ... 91630 26730 102-55-008 4013 I
A lot of empty values
Now, notice that with no NaN filter, columns 2, 3, 5, 6, 7 are all empty, and several others too further down. If you look at the actual data carefully, you will see that you have several consecutive tab characters, which means actual empty values in your data. That's just fine, presumably those fields were optional in the original database, but they're behind a lot of the NaNs that show up when not using na_filter=False.