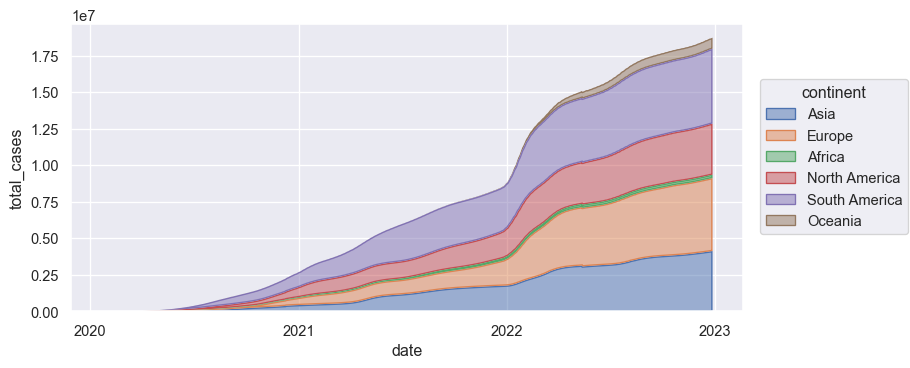
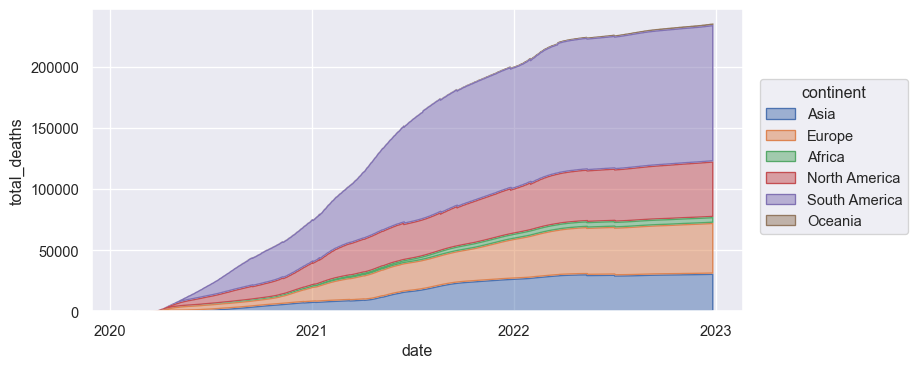
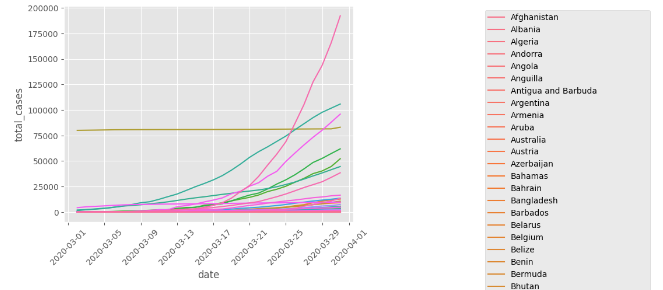
I have a dataset containing data on covid cases. 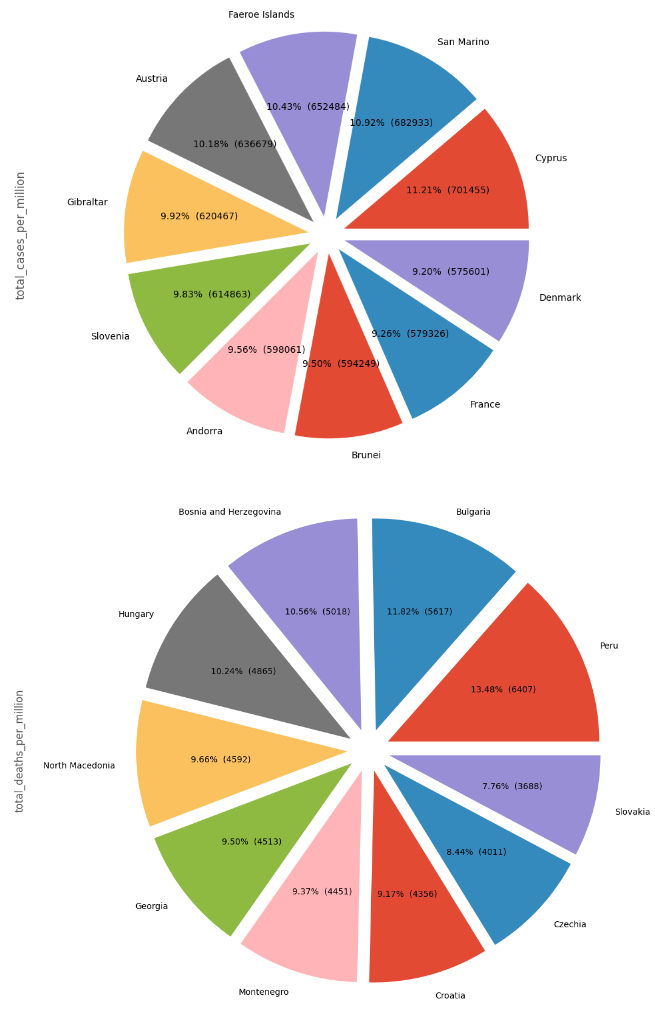
2:
total_cases_slice = df[['date', 'continent', 'total_cases']].dropna()
total_deaths_slice = df[['date', 'continent', 'total_deaths']].dropna()
s1 = so.Plot(total_cases_slice, x='date', y='total_cases', color='continent').add(so.Area(alpha=.5), so.Agg(), so.Stack()).layout(size=(8, 4))
s2 = so.Plot(total_deaths_slice, x='date', y='total_deaths', color='continent').add(so.Area(alpha=.5), so.Agg(), so.Stack()).layout(size=(8, 4))
s1.save('s1.png', bbox_inches='tight')
s2.save('s2.png', bbox_inches='tight')
3:
total_cases_march = df[df.date.gt('2020-03-01') & df.date.le('2020-03-31') & df.continent.notna()][['date', 'location', 'total_cases']]
s3 = sns.lineplot(data=total_cases_march, x='date', y='total_cases', hue='location')
plt.legend(bbox_to_anchor=(2.04, 1), loc="upper right")
for tick in s3.get_xticklabels():
tick.set_rotation(45)
plt.show()