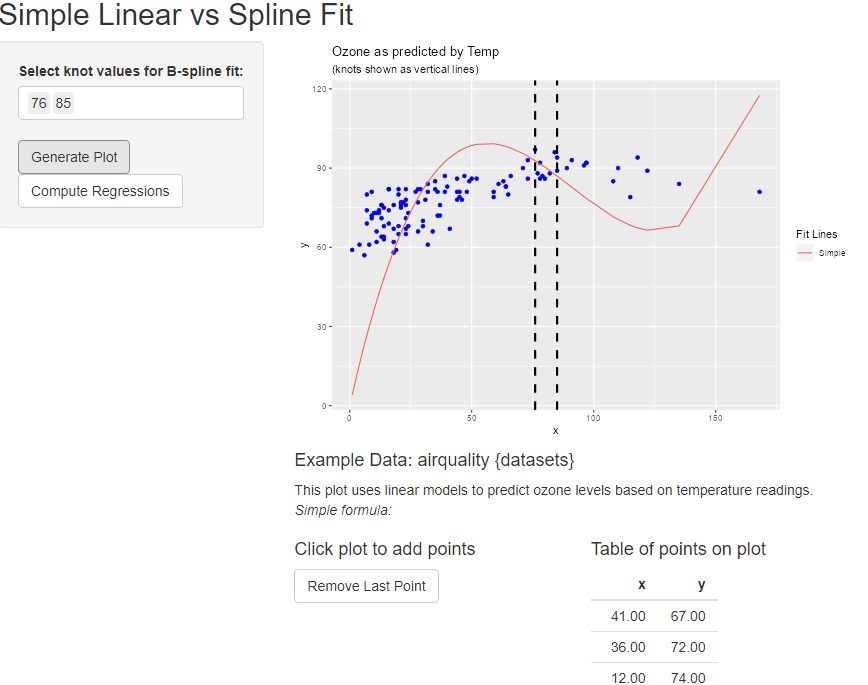
I have the following App:
The objective is to:
-
- Add new points to the plot when the user clicks on it.
-
- These are updated in the table (where you can remove points also)
-
- (Where the App fails): Plot the linear regression and spline regression based on the new users updated data.
When I comment-out the lines
#geom_line(aes(x=x, y=fitlm(), color="Simple"))
#geom_line(aes(x=x, y=fitbslm(), color="B-spline"))
in the ggplot renderPlot() function at the end, I am able to add points and update the plot without problem
The problem occurs when I try to add these two lines back into the plot and then the updated data is passed to the fitlm and fitBslm eventReactive() functions.
For some reason it doesn't want to re-compute the regressions and apply/update the plot.
Question:
How can I introduce the regressions to the ggplot based on the new users updated data. (I am happy with it updating automatically or through a button)
After clicking the Generate Plot button it makes the below plot. However, the plot failed Error: [object Object] when I click on the plot to add a new point.
App:
library(shiny)
library(dplyr)
library(splines2)
library(ggplot2)
# Get the Temp values, which defines the accepted range of knots
# for the b-spline model.
library(dplyr)
data("airquality")
airquality <- filter(airquality, !is.na(Ozone)) %>%
select(c(Ozone, Temp)) %>%
set_names(c("x", "y"))
uniqueTemps <- unique(airquality[order(airquality$x), "x"])
selectedTemps <- sample(uniqueTemps, 2)
# Define UI for application that draws a histogram
ui <- shinyUI(fluidPage(
# Application title
titlePanel("Simple Linear vs Spline Fit"),
# Sidebar
sidebarLayout(
sidebarPanel(
selectInput("knotSel", "Select knot values for B-spline fit:",
uniqueTemps, selected=selectedTemps,
multiple=TRUE),
actionButton("calcFit", "Generate Plot"),
actionButton("computeRegressions", "Compute Regressions")
),
# Show a plot of the generated distribution
mainPanel(
plotOutput("plot_splines", click = "plot_click"),
h4("Example Data: airquality {datasets}"),
p("This plot uses linear models to predict ozone levels based on temperature readings.",
tags$br(),
tags$em("Simple formula: ")
# tags$code("lm(Ozone ~ Temp I(Temp^2) I(Temp^3) - 1, airquality)"),
# tags$br(),
# tags$em("Spline formula: "),
# tags$code("lm(airquality$Ozone ~ bSpline(airquality$Temp, knots=getKnots(), degree=3) - 1)")
),
fluidRow(column(width = 6,
h4("Click plot to add points"),
actionButton("rem_point", "Remove Last Point")
#plotOutput("plot1", click = "plot_click")
),
column(width = 6,
h4("Table of points on plot"),
tableOutput("table"))),
fluidRow(column(width = 6,
DTOutput('tab1')),
column(width = 6,
DTOutput('tab2'))
)
)
)))
server <- (function(input, output) {
# Load the airquality dataset.
# data("airquality")
# # Remove observations lacking an Ozone measure.
# airquality <- filter(airquality, !is.na(Ozone)) %>%
# select(c(Ozone, Temp)) %>%
# set_names(c("y", "x"))
########################### Add selections to plot ###########################
## 1. set up reactive dataframe ##
values <- reactiveValues()
values$DT <- data.frame(x = numeric(),
y = numeric()
) %>%
bind_rows(airquality)
## 2. Create a plot ##
# output$plot1 = renderPlot({
# ggplot(values$DT, aes(x = x, y = y))
# geom_point(size = 5)
# lims(x = c(0, 100), y = c(0, 100))
# theme(legend.position = "bottom")
# # include so that colors don't change as more color/shape chosen
# # scale_color_discrete(drop = FALSE)
# # scale_shape_discrete(drop = FALSE)
# })
## 3. add new row to reactive dataframe upon clicking plot ##
observeEvent(input$plot_click, {
# each input is a factor so levels are consistent for plotting characteristics
add_row <- data.frame(x = input$plot_click$x,
y = input$plot_click$y
)
# add row to the data.frame
values$DT <- rbind(values$DT, add_row)
})
## 4. remove row on actionButton click ##
observeEvent(input$rem_point, {
rem_row <- values$DT[-nrow(values$DT), ]
values$DT <- rem_row
})
## 5. render a table of the growing dataframe ##
output$table <- renderTable({
values$DT
})
##############################################################################
# Fit the simple linear model
fitlm <- eventReactive(input$calcFit, {
slm <- lm(y ~ x I(x^2) I(x^3) - 1, values$DT)
fitlm <- slm$fitted.values
fitlm
})
# Get knot selection
getKnots <- reactive({as.integer(input$knotSel)})
# Fit the spline model, with the knot selection
fitBslm <- eventReactive(input$calcFit, {
bsMat <- bSpline(values$DT$x, knots=getKnots(), degree=3)
bslm <- lm(values$DT$y ~ bsMat - 1)
bslm
})
# observeEvent({
# print(fitBslm())
# })
# Generate the plot
output$plot_splines <- renderPlot({
splineMdl <- fitBslm()
fitbslm <- splineMdl$fitted.values
cols <- c("Simple"="#ef615c", "B-spline"="#20b2aa", "knot"="black")
g <- ggplot(values$DT, aes(x=x, y=y))
geom_point(color="blue")
geom_line(aes(x=x, y=fitlm(), color="Simple"))
#geom_line(aes(x=x, y=fitbslm(), color="B-spline"))
geom_vline(aes(color="knot"), xintercept=getKnots(), linetype="dashed", size=1)
scale_colour_manual(name="Fit Lines",values=cols)
ggtitle("Ozone as predicted by Temp", "(knots shown as vertical lines)")
g
})
output$tab1 <- renderDataTable(
airquality
)
output$tab2 <- renderDataTable(
values$DT
)
})
shinyApp(ui, server)
CodePudding user response:
Change eventReactive to just reactive. Also, you just need fitbslm in the second geom_line without (). Try this
server <- (function(input, output) {
########################### Add selections to plot ###########################
## 1. set up reactive dataframe ##
values <- reactiveValues()
values$DT <- data.frame(x = numeric(),
y = numeric()
) %>%
bind_rows(airquality)
## 3. add new row to reactive dataframe upon clicking plot ##
observeEvent(input$plot_click, {
# each input is a factor so levels are consistent for plotting characteristics
add_row <- data.frame(x = input$plot_click$x,
y = input$plot_click$y
)
# add row to the data.frame
values$DT <- rbind(values$DT, add_row)
})
## 4. remove row on actionButton click ##
observeEvent(input$rem_point, {
rem_row <- values$DT[-nrow(values$DT), ]
values$DT <- rem_row
})
## 5. render a table of the growing dataframe ##
output$table <- renderTable({
values$DT
})
##############################################################################
# Fit the simple linear model
# fitlm <- eventReactive(input$calcFit, {
fitlm <- reactive({
slm <- lm(y ~ x I(x^2) I(x^3) - 1, values$DT)
fitlm <- slm$fitted.values
fitlm
})
# Get knot selection
getKnots <- reactive({as.integer(input$knotSel)})
# Fit the spline model, with the knot selection
#fitBslm <- eventReactive(input$calcFit, {
fitBslm <- reactive({
bsMat <- bSpline(values$DT$x, knots=getKnots(), degree=3)
bslm <- lm(values$DT$y ~ bsMat - 1)
bslm
})
myPlot <- reactive({
splineMdl <- fitBslm()
fitbslm <- splineMdl$fitted.values
cols <- c("Simple"="#ef615c", "B-spline"="#20b2aa", "knot"="black")
g <- ggplot(values$DT, aes(x=x, y=y))
geom_point(color="blue")
geom_line(aes(x=x, y=fitlm(), color="Simple"))
geom_line(aes(x=x, y=fitbslm , color="B-spline"))
geom_vline(aes(color="knot"), xintercept=getKnots(), linetype="dashed", size=1)
scale_colour_manual(name="Fit Lines",values=cols)
ggtitle("Ozone as predicted by Temp", "(knots shown as vertical lines)")
g
})
# Generate the plot
output$plot_splines <- renderPlot({
myPlot()
})
output$tab1 <- renderDataTable(
airquality
)
output$tab2 <- renderDataTable(
values$DT
)
})