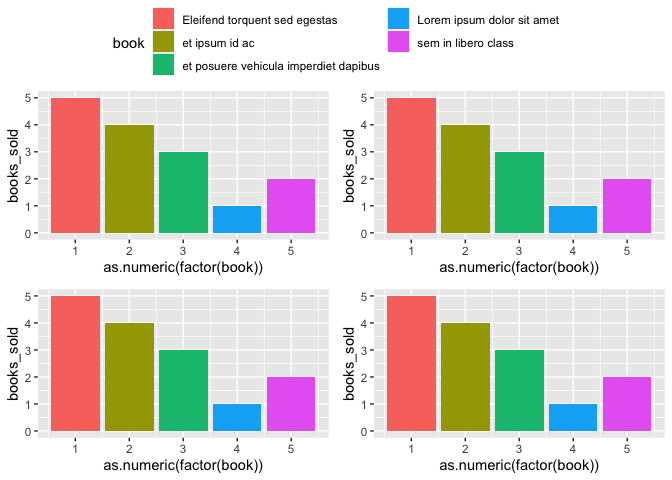
I have a problem with scale of legend, because text of legend crosses borders of plot. Any idea, how can I fix it? Split or resize?
Data cleaning:
Filtering data
df <- df %>%
filter(!(is.na(review)))
Changing state name to postal code
df <- df %>%
mutate(state = case_when(state == 'California' ~ 'CA',
state == 'Texas' ~ 'TX',
state == 'New York' ~ 'NY',
state == 'Florida' ~ 'FL',
TRUE ~ state))
Coding review column and adding new high_review
df <- df %>%
mutate(review = case_when(review == 'Poor' ~ 1,
review == 'Fair' ~ 2,
review == 'Good' ~ 3,
review == 'Great' ~ 4,
review == 'Excellent' ~ 5,),
high_review = ifelse(review >= 4, TRUE, FALSE))
Code of frames:
California
ca <- df %>%
filter(state == 'CA') %>%
group_by(book) %>%
summarize(books_sold = table(book)) %>%
arrange(-books_sold) %>%
mutate(rank = 1:5)
New York
ny <- df %>%
filter(state == 'NY') %>%
group_by(book) %>%
summarize(books_sold = table(book)) %>%
arrange(-books_sold) %>%
mutate(rank = 1:5)
Florida
fl <- df %>%
filter(state == 'FL') %>%
group_by(book) %>%
summarize(books_sold = table(book)) %>%
arrange(-books_sold) %>%
mutate(rank = 1:5)
Texas
tx <- df %>%
filter(state == 'TX') %>%
group_by(book) %>%
summarize(books_sold = table(book)) %>%
arrange(-books_sold) %>%
mutate(rank = 1:5)
This is how I created data frames for each state.
Code of plots:
ca_plot <- ggplot(data = ca, aes(x = reorder(rank, -books_sold), y = books_sold, fill = book))
geom_col()
geom_text(aes(label = books_sold), vjust = 1.1, size = 3)
ylab('Number of books sold')
xlab('Ranking')
ggtitle('California')
theme(legend.position = "none")
ny_plot <- ggplot(data = ny, aes(x = reorder(rank, -books_sold), y = books_sold, fill = book))
geom_col()
geom_text(aes(label = books_sold), vjust = 1.1, size = 3)
ylab('')
xlab('Ranking')
ggtitle('New York')
theme(legend.position = "none")
fl_plot <- ggplot(data = fl, aes(x = reorder(rank, -books_sold), y = books_sold, fill = book))
geom_col()
geom_text(aes(label = books_sold), vjust = 1.1, size = 3)
ylab('Number of books sold')
xlab('Ranking')
ggtitle('Florida')
theme(legend.position = "none")
tx_plot <- ggplot(data = tx, aes(x = reorder(rank, -books_sold), y = books_sold, fill = book))
geom_col()
geom_text(aes(label = books_sold), vjust = 1.1, size = 3)
ylab('')
xlab('Ranking')
ggtitle('Texas')
theme(legend.position = "none")
all_plot <- ggplot()
final_plot <- ggarrange(ca_plot, ny_plot, fl_plot, tx_plot, ncol = 2, nrow = 2,
common.legend = TRUE)
final_plot
And result:
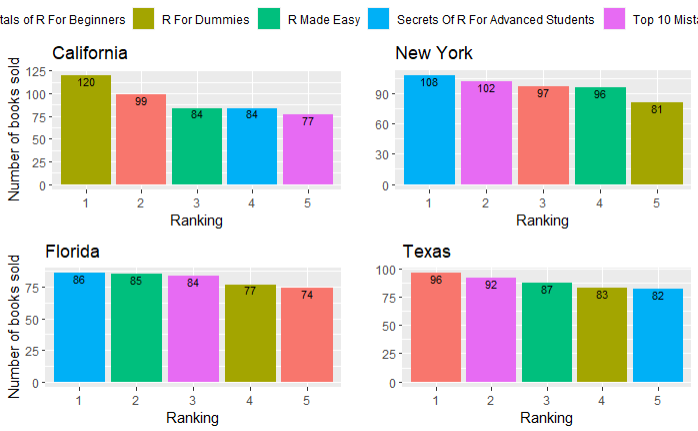
CodePudding user response:
As I suggested in my comment adding guides(fill = guide_legend(nrow = ...)) to one of your plots would be one option to split the legend into multiple rows.
As you provided no example data I created my own.
library(ggplot2)
library(ggpubr)
dat <- data.frame(
book <- c(
"Lorem ipsum dolor sit amet",
"sem in libero class",
"et posuere vehicula imperdiet dapibus",
"et ipsum id ac",
"Eleifend torquent sed egestas"
),
books_sold = 1:5
)
First, to reproduce your issue let's create some simple charts and use ggarrange to glue them together, which as you can see results in a legend clipped off at the plot margins because of the long legend text.
p1 <- p2 <- p3 <- p4 <- ggplot(dat, aes(as.numeric(factor(book)), books_sold, fill = book))
geom_col()
theme(legend.position = "none")
final_plot <- ggarrange(p1, p2, p3, p4,
ncol = 2, nrow = 2,
common.legend = TRUE
)
final_plot
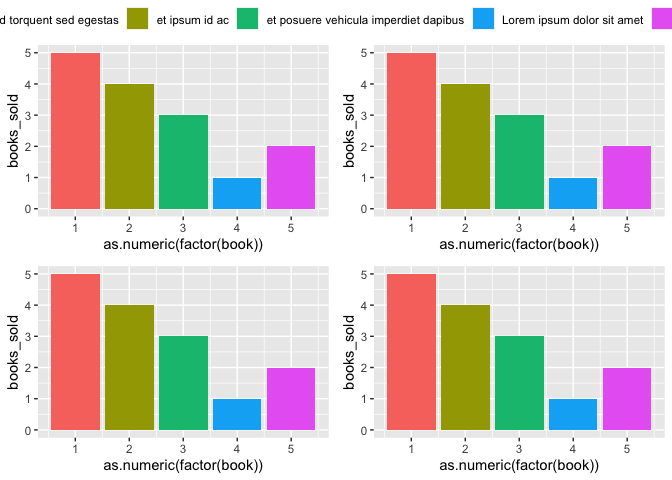
Now, adding guides(fill = guide_legend(nrow = 3)) to just one of your plots will split the legend into three rows:
p1 <- p1 guides(fill = guide_legend(nrow = 3))
final_plot <- ggarrange(p1, p2, p3, p4,
ncol = 2, nrow = 2,
common.legend = TRUE
)
final_plot